Abstract
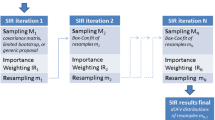
The Monte Carlo Parametric Expectation Maximization (MC-PEM) algorithm can approximate the true log-likelihood as precisely as needed and is efficiently parallelizable. Our objectives were to evaluate an importance sampling version of the MC-PEM algorithm for mechanistic models and to qualify the default estimation settings in SADAPT-TRAN. We assessed bias, imprecision and robustness of this algorithm in S-ADAPT for mechanistic models with up to 45 simultaneously estimated structural parameters, 14 differential equations, and 10 dependent variables (one drug concentration and nine pharmacodynamic effects). Simpler models comprising 15 parameters were estimated using three of the ten dependent variables. We set initial estimates to 0.1 or 10 times the true value and evaluated 30 bootstrap replicates with frequent or sparse sampling. Datasets comprised three dose levels with 16 subjects each. For simultaneous estimation of the full model, the ratio of estimated to true values for structural model parameters (median [5–95% percentile] over 45 parameters) was 1.01 [0.94–1.13] for means and 0.99 [0.68–1.39] for between-subject variances for frequent sampling and 1.02 [0.81–1.47] for means and 1.02 [0.47–2.56] for variances for sparse sampling. Imprecision was ≤25% for 43 of 45 means for frequent sampling. Bias and imprecision was well comparable for the full and simpler models. Parallelized estimation was 23-fold (6.9-fold) faster using 48 threads (eight threads) relative to one thread. The MC-PEM algorithm was robust and provided unbiased and adequately precise means and variances during simultaneous estimation of complex, mechanistic models in a 45 dimensional parameter space with rich or sparse data using poor initial estimates.




Similar content being viewed by others
Notes
The SADAPT-TRAN software is freely available via http://bmsr.usc.edu/.
References
Dempster AP, Laird N, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc B. 1977;39:1–38.
Schumitzky A. EM algorithms and two-stage methods in pharmacokinetic population analysis. In: D’Argenio DZ, editor. Advanced methods of pharmacokinetic and pharmacodynamic systems analysis. New York: Plenum Press; 1995. p. 145–60.
Walker S. An EM algorithm for nonlinear random effects models. Biometrics. 1996;52:934–44.
Bauer RJ, Guzy S, Ng C. A survey of population analysis methods and software for complex pharmacokinetic and pharmacodynamic models with examples. AAPS J. 2007;9:E60–83.
Wang X, Schumitzky A, D’Argenio DZ. Nonlinear random effects mixture models: maximum likelihood estimation via the EM algorithm. Comput Stat Data Anal. 2007;51:6614–23.
Wu C-FJ. On the convergence properties of the EM algorithm. Ann Stat. 1983;11:95–103.
Bustad A, Terziivanov D, Leary R, Port R, Schumitzky A, Jelliffe R. Parametric and nonparametric population methods: their comparative performance in analysing a clinical dataset and two Monte Carlo simulation studies. Clin Pharmacokinet. 2006;45:365–83.
Jelliffe RW. The USC*PACK PC programs for population pharmacokinetic modeling, modeling of large kinetic/dynamic systems, and adaptive control of drug dosage regimens. Proc Annu Symp Comput Appl Med Care. 1991: 922–4
Jelliffe RW, Schumitzky A, Bayard D, Milman M, Van Guilder M, Wang X, et al. Model-based, goal-oriented, individualised drug therapy. Linkage of population modelling, new ‘multiple model’ dosage design, Bayesian feedback and individualised target goals. Clin Pharmacokinet. 1998;34:57–77.
Schumitzky A. Nonparametric EM algorithms for estimating prior distributions. Appl Math Comput. 1991;45:143–57.
Laird N. Nonparametric maximum likelihood estimation of a mixing distribution. J Amer Stat Assn. 1978;73:805–11.
Bauer RJ. S-ADAPT/MCPEM user’s guide (Version 1.56). Software for Pharmacokinetic, Pharmacodynamic and Population Data Analysis. Berkeley, CA; 2008
D’Argenio DZ, Schumitzky A, Wang X. ADAPT 5 User’s Guide: Pharmacokinetic/Pharmacodynamic Systems Analysis Software. Biomedical Simulations Resource. Los Angeles, CA; 2009
D’Argenio DZ, Schumitzky A. A program package for simulation and parameter estimation in pharmacokinetic systems. Comput Programs Biomed. 1979;9:115–34.
Lavielle M. Monolix (Version 3.1)—A free software for the analysis of nonlinear mixed effects models. The Monolix Group. Paris; 2009
Beal SL, Sheiner LB, Boeckmann AJ, Bauer RJ. NONMEM user’s guides (1989–2009). Ellicott City, MD, USA: Icon Development Solutions; 2009.
Smith AFM, Gelfand AE. Bayesian statistics without tears: a sampling–resampling perspective. Am Stat. 1992;46:84–8.
Bauer RJ, Guzy S. Monte Carlo parametric expectation maximization (MC-PEM) method for analyzing population pharmacokinetic/pharmacodynamic data. In: D’Argenio DZ, editor. Advanced methods of pharmacokinetic and pharmacodynamic systems analysis. Boston, MA: Kluwer Academic Publishers; 2003. p. 135–63.
Bauer RJ. Technical guide on the population analysis methods in the S-ADAPT program (Version 1.56). Berkeley, CA; 2008
Bauer RJ. S-ADAPT/MCPEM Validation of Population Analysis of PK/PD Models. Berkeley, CA; 2006
Earp JC, Dubois DC, Molano DS, Pyszczynski NA, Keller CE, Almon RR, et al. Modeling corticosteroid effects in a rat model of rheumatoid arthritis I: mechanistic disease progression model for the time course of collagen-induced arthritis in Lewis rats. J Pharmacol Exp Ther. 2008;326:532–45.
Landersdorfer CB, DuBois DC, Almon RR, Jusko WJ. Mechanism-based modeling of nutritional and leptin influences on growth in normal and type 2 diabetic rats. J Pharmacol Exp Ther. 2009;328:644–51.
Ait-Oudhia S, Scherrmann JM, Krzyzanski W. Simultaneous pharmacokinetics/pharmacodynamics modeling of recombinant human erythropoietin upon multiple intravenous dosing in rats. J Pharmacol Exp Ther. 2010;334:897–910.
Bulitta JB, Yang JC, Yohonn L, Ly NS, Brown SV, D’Hondt RE, et al. Attenuation of colistin bactericidal activity by high inoculum of Pseudomonas aeruginosa characterized by a new mechanism-based population pharmacodynamic model. Antimicrob Agents Chemother. 2010;54:2051–62.
Mager DE, Jusko WJ. Development of translational pharmacokinetic–pharmacodynamic models. Clin Pharmacol Ther. 2008;83:909–12.
Bulitta JB, Ly NS, Yang JC, Forrest A, Jusko WJ, Tsuji BT. Development and qualification of a pharmacodynamic model for the pronounced inoculum effect of ceftazidime against Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009;53:46–56.
Ng CM, Joshi A, Dedrick RL, Garovoy MR, Bauer RJ. Pharmacokinetic–pharmacodynamic efficacy analysis of efalizumab in patients with moderate to severe psoriasis. Pharm Res. 2005;22:1088–100.
Ng CM, Patnaik A, Beeram M, Lin CC, Takimoto CH. Mechanism-based pharmacokinetic/pharmacodynamic model for troxacitabine-induced neutropenia in cancer patients. Cancer Chemother Pharmacol. 2010. doi:10.1007/s00280-010-1393-y.
Ng CM, Bai S, Takimoto CH, Tang MT, Tolcher AW. Mechanism-based receptor-binding model to describe the pharmacokinetic and pharmacodynamic of an anti-alpha(5)beta (1) integrin monoclonal antibody (volociximab) in cancer patients. Cancer Chemother Pharmacol. 2009;65:207–17.
Friberg LE, Karlsson MO. Mechanistic models for myelosuppression. Investig New Drugs. 2003;21:183–94.
Hooker AC, Ten Tije AJ, Carducci MA, Weber J, Garrett-Mayer E, Gelderblom H, et al. Population pharmacokinetic model for docetaxel in patients with varying degrees of liver function: incorporating cytochrome P4503A activity measurements. Clin Pharmacol Ther. 2008;84:111–8.
Bulitta JB, Zhao P, Arnold RD, Kessler DR, Daifuku R, Pratt J, et al. Mechanistic population pharmacokinetics of total and unbound paclitaxel for a new nanodroplet formulation versus Taxol in cancer patients. Cancer Chemother Pharmacol. 2009;63:1049–63.
Wiczling P, Lowe P, Pigeolet E, Ludicke F, Balser S, Krzyzanski W. Population pharmacokinetic modelling of filgrastim in healthy adults following intravenous and subcutaneous administrations. Clin Pharmacokinet. 2009;48:817–26.
Tanos PP, Isbister GK, Lalloo DG, Kirkpatrick CM, Duffull SB. A model for venom-induced consumptive coagulopathy in snake bite. Toxicon. 2008;52:769–80.
Wajima T, Isbister GK, Duffull SB. A comprehensive model for the humoral coagulation network in humans. Clin Pharmacol Ther. 2009;86:290–8.
Fung HL, Tsou PS, Bulitta JB, Tran DC, Page NA, Soda D, et al. Pharmacokinetics of 1, 4-butanediol in rats: bioactivation to gamma-hydroxybutyric acid, interaction with ethanol, and oral bioavailability. AAPS J. 2008;10:56–69.
Xu L, Eiseman JL, Egorin MJ, D’Argenio DZ. Physiologically-based pharmacokinetics and molecular pharmacodynamics of 17-(allylamino)-17-demethoxygeldanamycin and its active metabolite in tumor-bearing mice. J Pharmacokinet Pharmacodyn. 2003;30:185–219.
Marathe A, Peterson MC, Mager DE. Integrated cellular bone homeostasis model for denosumab pharmacodynamics in multiple myeloma patients. J Pharmacol Exp Ther. 2008;326:555–62.
Beal SL. Ways to fit a PK model with some data below the quantification limit. J Pharmacokinet Pharmacodyn. 2001;28:481–504.
Sheiner LB, Beal SL. Evaluation of methods for estimating population pharmacokinetics parameters. I. Michaelis–Menten model: routine clinical pharmacokinetic data. J Pharmacokinet Biopharm. 1980;8:553–71.
Sheiner LB, Beal SL. Evaluation of methods for estimating population pharmacokinetic parameters II. Biexponential model and experimental pharmacokinetic data. J Pharmacokinet Biopharm. 1981;9:635–51.
Steimer JL, Mallet A, Golmard JL, Boisvieux JF. Alternative approaches to estimation of population pharmacokinetic parameters: comparison with the nonlinear mixed-effect model. Drug Metab Rev. 1984;15:265–92.
Roe DJ. Comparison of population pharmacokinetic modeling methods using simulated data: results from the Population Modeling Workgroup. Stat Med. 1997;16:1241–57. discussion 57–62.
Duffull SB, Kirkpatrick CM, Green B, Holford NH. Analysis of population pharmacokinetic data using NONMEM and WinBUGS. J Biopharm Stat. 2005;15:53–73.
Jonsson S, Kjellsson MC, Karlsson MO. Estimating bias in population parameters for some models for repeated measures ordinal data using NONMEM and NLMIXED. J Pharmacokinet Pharmacodyn. 2004;31:299–320.
Plan EL, Maloney A, Troconiz IF, Karlsson MO. Performance in population models for count data, part I: maximum likelihood approximations. J Pharmacokinet Pharmacodyn. 2009;36:353–66.
Jonsson EN, Wade JR, Karlsson MO. Nonlinearity detection: advantages of nonlinear mixed-effects modeling. AAPS PharmSci. 2000;2:E32.
Krzyzanski W, Jusko WJ. Note: caution in use of empirical equations for pharmacodynamic indirect response models. J Pharmacokinet Biopharm. 1998;26:735–41.
Krzyzanski W, Jusko WJ. Indirect pharmacodynamic models for responses with multicompartmental distribution or polyexponential disposition. J Pharmacokinet Pharmacodyn. 2001;28:57–78.
Petzold LR. Automatic selection of methods for solving stiff and nonstiff systems of ordinary diferential equations. Siam J Sci Stat Comput. 1983;4:136–48.
D’Argenio DZ, Schumitzky A. ADAPT II user’s guide: pharmacokinetic/pharmacodynamic systems analysis software. Biomedical Simulations Resource. Los Angeles; 2006
Delyon B, Lavielle M, Moulines E. Convergence of a stochastic approximation version of the EM algorithm. Ann Stat. 1999;27:94–128.
Kuhn E, Lavielle M. Coupling a stochastic approximation version of EM with a MCMC procedure. ESAIM Probab Stat. 2004;8:115–31.
Acknowledgment
We greatly thank Drs. David Z. D’Argenio, Robert J. Bauer, Nicholas H. G. Holford, Alan Schumitzky, and Chee M. Ng for insightful discussions on EM algorithms and valuable comments for the design and functionality of SADAPT-TRAN. We thank Dr. William J. Jusko for providing a stimulating environment during our post-doctoral fellowships at the University at Buffalo. We thank the three reviewers of this manuscript for very helpful comments. The Perl scripts included in the SADAPT-TRAN package were completely written by JBB since 2003 and do not contain any code from any other software. The work presented here was not supported by a funding source.
Conflicts of Interest
None for all authors.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bulitta, J.B., Landersdorfer, C.B. Performance and Robustness of the Monte Carlo Importance Sampling Algorithm Using Parallelized S-ADAPT for Basic and Complex Mechanistic Models. AAPS J 13, 212–226 (2011). https://doi.org/10.1208/s12248-011-9258-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1208/s12248-011-9258-9