Abstract
Background
Assisted reproductive technologies are associated with altered methylation in term placenta. However, it is unclear whether methylation patterns are the result of fertility treatments or intrauterine environment. Thus, we set out to determine whether there are differences in the first-trimester placenta that may be altered by the underlying fertility treatments. Genome-wide DNA methylation analyses from chorionic villus sampling (CVS) from matched singleton pregnancies conceived using in vitro fertilization (IVF), non-IVF fertility treatment (NIFT), or those conceived spontaneously were performed using Illumina Infinium HumanMethylation450 BeadChip from 15 matched CVS samples. Nanofluidic quantitative polymerase chain reaction (qPCR) of differently methylated genes was performed in a confirmatory cohort of 23 IVF conceptions and 24 NIFT conceptions.
Results
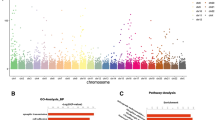
Global methylation was similar among the IVF, NIFT, and spontaneous conceptions. However, differential methylation from IVF and NIFT pregnancies was present at 34 CpG sites, which was significantly different. Of those, 14 corresponded to known genes, with methylation changes detected at multiple loci in 3 genes, anaphase-promoting complex subunit 2 (ANAPC2), C-X-C motif chemokine ligand 14 (CXCL14), and regulating synaptic membrane exocytosis 1 (RIMS1). Nanofluidic qPCR of differentially methylated genes identified pre T-cell antigen receptor alpha (PTCRA) to be significantly downregulated in IVF versus NIFT conceptions.
Conclusion
Although global methylation patterns are similar, there are differences in methylation of specific genes in IVF compared to NIFT conceptions, leading to altered gene expression. PTCRA was differentially methylated and downregulated in IVF conceptions, warranting further investigation. It remains to be determined whether these changes affect placentation and whether it is due to the more profound underlying infertility requiring IVF, yet these data provide unique insight into the first-trimester placental epigenome.
Similar content being viewed by others
References
Katari S, Turan N, Bibikova M, et al. DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum Mol Genet. 2009;18(20):3769–3778.
Gosden R, Trasler J, Lucifero D, Faddy M. Rare congenital dis-orders, imprinted genes, and assisted reproductive technology. Lancet. 2003;361(9373):1975–1977.
Sunderam S, Chang J, Flowers L, et al., Assisted reproductive technology surveillance—United States, 2006. MMWR Surveill Summ. 2009;58(5):1–25.
Hansen M, Kurinczuk JJ, Bower C, Webb S. The risk of major birth defects after intracytoplasmic sperm injection and in vitro fertilization. N Engl J Med. 2002;346(10):725–730.
Klemetti R, Gissler M, Sevón T, Koivurova S, Ritvanen A, Hemminki E. Children born after assisted fertilization have an increased rate of major congenital anomalies. Fertil Steril. 2005;84(5):1300–1307.
Schieve LA, Meikle SF, Ferre C, Peterson HB, Jeng G, Wilcox LS. Low and very low birth weight in infants conceived with use of assisted reproductive technology. N Engl J Med. 2002;346(10):731–737.
Strömberg B, Dahlquist G, Ericson A, Finnström O, Köster M, Stjernqvist K. Neurological sequelae in children born after in-vitro fertilisation: a population-based study. Lancet. 2002;359(9305):461–465.
Manipalviratn S, DeCherney A, Segars J. Imprinting disorders and assisted reproductive technology. Fertil Steril. 2009;91(2):305–315.
Vermeiden JP, Bernardus RE. Are imprinting disorders more pre-valent after human in vitro fertilization or intracytoplasmic sperm injection? Fertil Steril. 2013;99(3):642–651.
Feinberg AP. Genome-scale approaches to the epigenetics of common human disease. Virchows Arch. 2010;456(1):13–21.
Feinberg AP, Irizarry RA, Fradin D, et al. Personalized epige-nomic signatures that are stable over time and covary with body mass index. Sci Transi Med. 2010;2(49):49ra67.
Monk M, Salpekar A. Expression of imprinted genes in human preimplantation development. Mol Cell Endocrinol. 2001; 183(suppl 1):S35–S40.
Jirtle RL, Sander M, Barrett JC. Genomic imprinting and envi-ronmental disease susceptibility. Environ Health Perspect. 2000;108(3):271–278.
Zaitseva I, Zaitsev S, Alenina N, Bader M, Krivokharchenko A. Dynamics of DNA-demethylation in early mouse and rat embryos developed in vivo and in vitro. Mol Reprod Dev. 2007;74(10):1255–1261.
Doherty AS, Mann MR, Tremblay KD, Bartolomei MS, Schultz RM. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol Reprod. 2000;62(6):1526–1535.
Khosla S, Dean W, Brown D, Reik W, Feil R. Culture of preim-plantation mouse embryos affects fetal development and the expression of imprinted genes. Biol Reprod. 2001;64(3):918–926.
El Hajj N, Haaf T. Epigenetic disturbances in in vitro cultured gametes and embryos: implications for human assisted reproduc-tion. Fertil Steril. 2013;99(3):632–641.
Al-Khtib M, Blachère T, Guérin JF, Lefèvre A. Methylation pro-file of the promoters of Nanog and Oct4 in ICSI human embryos. Hum Reprod. 2012;27(10):2948–2954.
Khoueiry R, Ibala-Romdhane S, Al-Khtib M, et al. Abnormal methylation of KCNQIOTI and differential methylation of H19 imprinting control regions in human ICSI embryos. Zygote. 2013;21(2):129–138.
Nelissen EC, Dumoulin JC, Daunay A, Evers JL, Tost J, van Montfoort AP. Placentas from pregnancies conceived by IVF/ICSI have a reduced DNA methylation level at the H19 and MEST differentially methylated regions. Hum Reprod. 2013;28(4):1117–1126.
Li L, Wang L, Le F, et al., Evaluation of DNA methylation status at differentially methylated regions in IVF-conceived newborn twins. Fertil Steril. 2011;95(6):1975–1979.
Rancourt RC, Harris HR, Michels KB. Methylation levels at imprinting control regions are not altered with ovulation induction or in vitro fertilization in a birth cohort. Hum Reprod. 2012;27(7):2208–2216.
Oliver VF, Miles HL, Cutfield WS, Hofman PL, Ludgate JL, Morison IM. Defects in imprinting and genome-wide DNA methylation are not common in the in vitro fertilization population. Fertil Steril. 2012;97(1):147–153.e7.
Wong EC, Hatakeyama C, Robinson WP, Ma S. DNA methyla-tion at H19/IGF2 ICR1 in the placenta of pregnancies conceived by in vitro fertilization and intracytoplasmic sperm injection. Fertil Steril. 2011;95(8):2524–2526.el-e3.
Melamed N, Choufani S, Wilkins-Haug LE, Koren G, Weksberg R. Comparison of genome-wide and gene-specific DNA methyla-tion between ART and naturally conceived pregnancies. Epige-netics. 2015;10(6):474–483.
Buckberry S, Bianco-Miotto T, Hiendleder S, Roberts CT. Quan-titative allele-specific expression and DNA methylation analysis of HI 9, IGF2 and IGF2R in the human placenta across gestation reveals H19 imprinting plasticity. PLoS One. 2012;7(12):e51210.
Herzog E, Galvez J, Roks A. Tissue-specific DNA methylation profiles in newborns. Clin Epigenetics. 2013;5(1):8.
Bird LM. Angelman syndrome: review of clinical and molecular aspects. Appl Clin Genet. 2014;7:93–104.
Niu Y, Wang Z, Huang H, et al. Activated pregnane X receptor inhibits cervical cancer cell proliferation and tumorigenicity by inducing G2/M cell-cycle arrest. Cancer Lett. 2014;347(1):88–97.
Kuang H, Chen Q, Zhang Y. The cytokine gene CXCL14 restricts human trophoblast cell invasion by suppressing gelatinase activity. Endocrinology. 2009;150(12):5596–5605.
Cheong CY, Chng K, Lim MK, et al. Alterations to DNA methy-lation and expression of CXCL14 are associated with suboptimal birth outcomes. J Hum Genet. 2014;59(9):504–511.
Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41(2):178–186.
Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G. Enhancers: five essential questions. Nat Rev Genet. 2013;14(4):288–295.
Lavrov AV, Chelysheva EY, Smirnikhina SA, et al. Frequent variations in cancer-related genes may play prognostic role in treatment of patients with chronic myeloid leukemia. BMC Genet. 2016;17(suppl 1):14.
Rodriguez RM, Suarez-Alvarez B, Mosén-Ansorena D, et al. Regulation of the transcriptional program by DNA methylation during human alphabeta T-cell development. Nucleic Acids Res. 2015;43(2):760–774.
Brandeis MI, Frank D, Keshet I, et al. Spl elements protect a CpG island from de novo methylation. Nature. 1994;371(6496):435–438.
Macleod D, Charlton J, Mullins J, Bird AP. Spl sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8(19):2282–2292.
Smale ST. Pioneer factors in embryonic stem cells and differen-tiation. Curr Opin Genet Dev. 2010;20(5):519–526.
Brameld KJ, Dickinson JE, O’Leary P, et al. First trimester pre-dictors of adverse pregnancy outcomes. Aust N Z J Obstet Gynaecol. 2008;48(6):529–535.
Jelliffe-Pawlowski LL, Shaw GM, Currier RJ, et al. Association of early-preterm birth with abnormal levels of routinely collected first- and second-trimester biomarkers. Am J Obstet Gynecol. 2013;208(6):492.e1–e11.
Hu XL, Feng C, Lin XH, et al. High maternal serum estradiol environment in the first trimester is associated with the increased risk of small-for-gestational-age birth. J Clin Endocrinol Metab. 2014;99(6):2217–2224.
Smith GC, Smith MF, McNay MB, Fleming JE. First-trimester growth and the risk of low birth weight. N Engl J Med. 1998;339(25):1817–1822.
Chu T, Handley D, Bunce K, Surti U, Hogge WA, Peters DG. Structural and regulatory characterization of the placental epigenome at its maternal interface. PLoS One. 2011;6(2):e14723.
Banister CE, Koestler DC, Maccani MA, Padbury JF, Houseman EA, Marsit CJ. Infant growth restriction is associated with distinct patterns of DNA methylation in human placentas. Epigenetics. 2011;6(7):920–927.
Anton L, Brown AG, Bartolomei MS, Elovitz MA. Differential methylation of genes associated with cell adhesion in preeclamptic placentas. PLoS One. 2014;9(6):e100148.
Song S, Ghosh J, Mainigi M, et al. DNA methylation differences between in vitro- and in vivo-conceived children are associated with ART procedures rather than infertility. Clin Epigenetics. 2015;7(1):41.
El Hajj N, Schneider E, Lehnen H, Haaf T. Epigenetics and life-long consequences of an adverse nutritional and diabetic intrau-terine environment. Reproduction. 2014;148(6):R111–R120.
Founds SA, Conley YP, Lyons-Weiler JF, Jeyabalan A, Hogge WA, Conrad KP. Altered global gene expression in first trimester placentas of women destined to develop preeclampsia. Placenta. 2009;30(1):15–24.
Conway DA, Liem J, Patel S, Fan KJ, Williams J III, Pisarska MD. The effect of infertility and assisted reproduction on first-trimester placental and fetal development. Fertil Steril. 2011;95(5):1801–1804.
Jackson S, Hong C, Wang ET, Alexander C, Gregory KD, Pisarska MD. Pregnancy outcomes in very advanced maternal age pregnancies: the impact of assisted reproductive technology. Fertil Steril. 2015;103(1):76–80.
Chandra A, Copen CE, Stephen EH. Infertility service use in the United States: data from the National Survey of Family Growth, 1982–2010. Natl Health Stat Report. 2014;(73):1–21.
Huang A, Adusumalli J, Patel S, Liem J, Williams J III, Pisarska MD. Prevalence of chromosomal mosaicism in pregnancies from couples with infertility. Fertil Steril. 2009;91(6):2355–2360.
Conway DA, Patel SS, Liem J, et al. The risk of cytogenetic abnormalities in the late first trimester of pregnancies conceived through assisted reproduction. Fertil Steril. 2011;95(2):503–506.
Clifton VL. Review: Sex and the human placenta: mediating dif-ferential strategies of fetal growth and survival. Placenta. 2010; 31(suppl):S33–S39.
Morris TJ, Butcher LM, Feber A, et al. ChAMP: 450k Chip Analysis Methylation Pipeline. Bioinformatics. 2014;30(3):428–430.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B (Methodol). 1995;57(1):289–300.
Kalisky T, Quake SR. Single-cell genomics. Nat Methods. 2011;8(4):311–314.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xu, N., Barlow, G.M., Cui, J. et al. Comparison of Genome-Wide and Gene-Specific DNA Methylation Profiling in First-Trimester Chorionic Villi From Pregnancies Conceived With Infertility Treatments. Reprod. Sci. 24, 996–1004 (2017). https://doi.org/10.1177/1933719116675056
Published:
Issue Date:
DOI: https://doi.org/10.1177/1933719116675056