Abstract
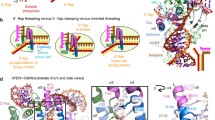
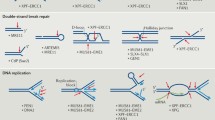
Flap endonuclease 1 (FEN1) is strongly specific for the substrate structure. The natural substrates of FEN1 are 5′-flap structures formed by three DNA strands one of which has an unpaired 5′ portion (a flap). Flap structures occur as intermediates in various processes of DNA metabolism such as DNA replication, Okazaki fragment maturation during replication of the lagging strand, and strand displacement DNA synthesis in base excision repair. In addition, FEN1 possesses 5′-exonuclease activity and gap endonuclease activity, discovered recently. FEN1 physically and functionally interacts with many DNA replication and repair proteins, including the proliferating cell nuclear antigen, helicase/nuclease Dna2, WRN, BLM, replication protein A, apurinic/apyrimidinic endonuclease 1, DNA polymerase β, poly(ADP-ribose) polymerase 1, high mobility group protein 1, human immunodeficiency virus integrase, transcriptional coactivator p300, chromatin proteins, cyclin-dependent kinases (Cdk1 and Cdk2), and cyclin A. FEN1 activity is important for maintaining the integrity of repetitive sequences in the genome. Recent data suggest a correlation between defects in hFEN1 activity and the development and progression of neurodegenerative diseases and cancer. Hence, FEN1 exerts a dramatic effect on cell growth and development and, consequently, attracts particular interest.
Similar content being viewed by others
References
Lee B., Nguyen L.H., Barsky D., Fernandes M., Wilson D.M. III. 1999. Molecular interactions of human Exo1 with DNA. Nucleic Acids Res. 30, 942–949.
Lieber M.R. 1997. The FEN1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays. 19, 233–240.
Shen B., Singh P., Liu R., Qiu J., Zheng L., Finger L.D., Alas S. 2005. Multiple but dissectible functions of FEN-1 nucleases in nucleic acid processing, genome stability and diseases. Bioessays. 27, 717–729.
Liu Y., Kao H.I., Bambara R.A. 2004. Flap endonuclease 1: A central component of DNA metabolism. Annu. Rev. Biochem. 73, 589–615.
Henneke G., Friedrich-Heineken E., Hübscher U. 2003. Flap endonuclease 1: A novel tumor suppressor protein. Trends Biochem. Sci. 28, 384–390.
Harrington J.J., Lieber M.R. 1994. The characterization of a mammalian structure-specific endonuclease. EMBO J. 13, 1235–1246.
Negritto M.C., Qiu J., Ratay D.O., Shen B., Bailis, A.M. 2001. Novel function of Rad27 (FEN-1) in restricting short-sequence recombination. Mol. Cell. Biol. 21, 2349–2358.
Wu X., Li J., Hsieh C., Burgers P.M.J., Lieber M.R. 1996. Processing of branched DNA intermediates by a complex of human FEN-1 and PCNA. Nucleic Acids Res. 24, 2036–2043.
Murante R.S., Rust L., Bambara R.A. 1995. Calf 5′ to 3′ exo/endonuclease must slide from a 5′ end of the substrate to perform structure-specific cleavage. J. Biol. Chem. 270, 30377–30383.
Harrington J.J., Lieber M.R. 1995. DNA structural elements required for FEN-1 binding. J. Biol. Chem. 270, 4503–4508.
Nazarkina Zh.K., Pyshnyi D.V., Pyshnaya I.A., Lavrik O.I., Khodyreva S.N. 2005. Use of modified flap structures for study of base excision repair proteins. Biokhimiya. 70, 1613–1622.
Erzberger J., Wilson D. III. The role of Mg2+ and specific amino acid residues in the catalytic reaction of the major human abasic endonuclease: New insights from EDTA-resistant incision of acyclic abasic site analogs and site-directed mutagenesis. J. Mol. Biol. 290, 447–457.
Takeshita M., Chang C.N., Johnson F., Will S., Grollman A.P. 1987. Oligodeoxynucleotides containing synthetic abasic sites: Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J. Biol. Chem. 262, 10171–10179.
Nazarkina Zh.K., Petrousseva I.O, Safronov I.V, Lavrik O.I, Khodyreva S.N. 2003. Interaction of flap endonuclease-1 and replication protein A with photo-reactive intermediates of DNA repair. Biokhimiya. 68, 1138–1148.
Kaiser M.W., Lyamicheva N., Ma W., Miller C., Neri B., Fors L., Lyamichev V.I. 1999. A comparison of eubacterial and archaeal structure-specific 5′-exonucleases. J. Biol. Chem. 274, 21387–21394.
Kao H.I., Henricksen L.A., Liu Y., Bambara R.A. 2002. Cleavage specificity of Saccharomyces cerevisiae flap endonuclease 1 suggests a double-flap structure as the cellular substrate. J. Biol. Chem. 277, 14379–14389.
Friedrich-Heineken E., Henneke G., Ferrari E., Hubscher U. 2003. The acetylatable lysines of human Fen1 are important for endo-and exonuclease activities. J. Mol. Biol. 328, 73–84.
Storici F., Henneke G., Ferrari E., Gordenin D., Hubscher U., Resnick M. 2002. The acetylatable lysines of human Fen1 are important for endo-and exonuclease activities. EMBO J. 21, 5930–5942.
Harrington C., Perrino F.W. 1995. The effects of cytosine arabinoside on RNA-primed DNA synthesis by DNA polymerase α-primase. J. Biol. Chem. 270, 26664–26669.
Khlimankov D.Yu., Rechkunova N.I., Kolpashchikov D.M., Khodyreva S.N., Lavrik O.I. 2001. Affinity labeling of flap endonuclease FEN-1 by photoreactive DNAs. Biokhimiya. 66, 905–912.
Zheng L., Zhou M., Chai Q., Parrish J., Xue D., Patrick S.M., Turchi J.J., Yannone S.M., Chen D., Shen B. 2005. Novel function of the flap endonuclease 1 complex in processing stalled DNA replication forks. EMBO Rep. 6, 83–89.
Liu R., Qiu J., Finger L.D., Zheng L., Shen B. 2006. The DNA-protein interaction modes of FEN-1 with gap substrates and their implication in preventing duplication mutations. Nucleic Acids Res. 34, 1772–1784.
Hohl M., Dunand-Sauthier I., Staresincic L., Jaquier-Gubler P., Thorel F., Modesti M., Clarkson S.G., Scharer O.D. 2007. Domain swapping between FEN-1 and XPG defines regions in XPG that mediate nucleotide excision repair activity and substrate specificity. Nucleic Acids Res. 35, 3053–3063.
Hosfield D.J., Mol C.D., Shen B., Tainer J.A. 1998. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: Coupling DNA and PCNA binding to FEN-1 activity. Cell. 95, 135–146.
Bornarth C.J., Ranalli T.A., Henrickesen L.A., Wahl A.F., Bambara R.A. 1999. Effect of flap modifications on human FEN1 cleavage. Biochemistry. 38, 13347–13354.
Hwang K.Y., Baek K., Kim H.Y., Cho Y. 1998. The crystal structure of flap endonuclease-1 from Methanococcus jannaschii. Nature Struct. Biol. 5, 707–713.
Dervan J., Feng M., Patel D., Grasby J., Artymiuk P., Ceska T.A., Sayers J.R. 2002. Interactions of mutant and wild-type flap endonucleases with oligonucleotide substrates suggest an alternative model of DNA binding. Proc. Natl. Acad. Sci. USA. 99, 8542–8547.
Chapados B.R., Hosfeld D.J., Han S., Qiu J., Yelent B., Shen B., Tainer J.A. 2004. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 116, 39–50.
Friedrich-Heineken E., Hubscher U. 2004. The Fen1 extrahelical 3′-flap pocket is conserved from archaea to human and regulates DNA substrate specificity. Nucleic Acids Res. 32, 2520–2528.
Tom S., Henricksen L.A., Bambara R.A. 2000. Mechanism whereby proliferating cell nuclear antigen (PCNA) stimulates endonuclease-1. J. Biol. Chem. 275, 10498–10505.
Li X., Li J., Harrington J., Lieber M.R., Burgers P.M. 1995. Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J. Biol. Chem. 270, 22109–22112.
Chen J.J., Chen S., Saha P., Dutta A. 1996. p21Cip1/Waf1 disrupts the recruitment of human Fen1 by proliferating-cell nuclear antigen into the DNA replication complex. Proc. Natl. Acad. Sci. USA. 93, 11597–11602.
Budd M.E., Campbell J.L. 1997. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol. Cell. Biol. 17, 2136–2142.
Stewart J.A., Campbell J.L., Bambara R.A. 2006. Flap endonuclease disengages Dna2 helicase/nuclease from Okazaki fragment flaps. J. Biol. Chem. 281, 38565–38572.
Brosh R.M.J., von Kobbe C., Sommers J.A., Karmakar P., Opresko P.L., Piotrowski J., Dianova I., Dianov G.L., Bohr V.A. 2001. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 20, 5791–5801.
Brosh R.M.J., Driscoll H.C., Dianov G.L., Sommers J.A. 2002. Biochemical characterization of the WRN-FEN-1 functional interaction. Biochemistry. 41, 12204–12216.
Sharma S., Sommers J.A., Wu L., Bohr V.A., Hickson I.D., Brosh R.M.J. 2004. Stimulation of flap endonuclease-1 by the Bloom’s syndrome protein. J. Biol. Chem. 279, 9847–9856.
Sharma S., Otterlei M., Sommers J.A., Driscoll H.C., Dianov G.L., Kao H.I., Bambara R.A., Brosh R.M.J. 2004. WRN helicase and FEN-1 form a complex upon replication arrest and together process branch migrating DNA structures associated with the replication fork. Mol. Biol. Cell. 15, 734–750.
Wang W., Bambara R.A. 2005. Human Bloom protein stimulates flap endonuclease 1 activity by resolving DNA secondary structure. J. Biol. Chem. 280, 5391–5399.
Bae S.H., Bae K.H., Kim J.A., Seo Y.S. 2001. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 412, 456–461.
Kao H.I., Veeraraghavan J., Polaczek P., Campbell J.L., Bambara R.A. 2004. On the roles of Saccharomyces cerevisiae Dna2p and Flap endonuclease 1 in Okazaki fragment processing. J. Biol. Chem. 279, 15014–15024.
Dianova I., Bohr V.A., Dianov G.L. 2001. Interaction of human AP endonuclease 1 with flap endonuclease 1 and proliferating cell nuclear antigen involved in long-patch base excision repair. Biochemistry. 40, 12639–12644.
Ranalli T.A., Tom S., Bambara R.A. 2002. AP endonuclease 1 coordinates flap endonuclease 1 and DNA ligase I activity in long patch base excision repair. J. Biol. Chem. 277, 41715–41724.
Prasad R., Dianov G.L., Bohr V.A., Wilson S.H. 2000. FEN1 stimulation of DNA polimerase β mediates an excision step in mammalian long patch base excision repair. J. Biol. Chem. 6, 4460–4466.
Prasad R., Lavrik O.I., Kim S.J., Kedar P., Yang X.P., vande Berg B.J., Wilson S.H. 2001. DNA polymerase β-mediated long patch base excision repair: Poly(ADP-ribose) polymerase-1 stimulates strand displacement DNA synthesis. J. Biol. Chem. 35, 32411–32414.
Sukhanova M.V., Khodyreva S.N., Lavrik O.I. 2006. Influence of poly(ADP-ribose) polymerase-1 and its apoptotoic 24-kDa fragment on repair of DNA duplexes in bovine testis nuclear extract. Biokhimiya. 71, 909–923.
Prasad R., Liu Y., Deterding L.J., Poltoratsky V.P., Kedar P.S., Horton J.K., Kanno S.-I., Asagoshi K., Hou E.W., Khodyreva S.N., Lavrik O.I., Tomer K.B., Yasui A., Wilson S.H. 2007. HMGB1 is a cofactor in mammalian base excision repair. Mol. Cell. 27, 829–841.
Faust E.A., Triller H. 2002. Stimulation of human flap endonuclease 1 by human immunodeficiency virus type 1 integrase: Possible role for flap endonuclease 1 in 5′-end processing of human immunodeficiency virus type 1 integration intermediates. J. Biomed. Sci. 9, 273–287.
Hasan S., Stucki M., Hassa P.O., Imhof R., Gehrig P., Hunziker P., Hubscher U., Hottiger M.O. 2001. Regulation of human flap endonuclease-1 activity by acetylation through the transcriptional coactivator p300. Mol. Cell. 7, 1221–1231.
Huggins C.F., Chafin D.R., Aoyagi S., Henricksen L.A., Bambara R.A., Hayes J.J. 2002. Flap endonuclease 1 efficiently cleaves base excision repair and DNA replication intermediates assembled into nucleosomes. Mol. Cell. 10, 1201–1211.
Henneke G., Koundrioukoff S., Hubscher U. 2003. Phosphorylation of human Fen1 by cyclin-dependent kinase modulates its role in replication fork regulation. Oncogene. 22, 4301–4313.
Parrish J.Z., Yang C.L., Shen B.H., Xue D. 2003. CRN-1, a Caenorhabditis elegans FEN-1 homologue, cooperates with CPS-6/EndoG to promote apoptotic DNA degradation. EMBO J. 22, 3451–3460.
Friedrich-Heineken E., Toueille M., Tannler B., Burki C., Ferrari E., Hottiger M.O., Hubscher U. 2005. The two DNA clamps Rad9/Rad1/Hus1 complex and proliferating cell nuclear antigen differentially regulate flap endonuclease 1 activity. J. Mol. Biol. 353, 980–989.
Chai Q., Zheng L., Zhou M., Turchi J.J., Shen B. 2003. Interaction and stimulation of human FEN-1 nuclease activities by heterogeneous nuclear ribonucleoprotein A1 in R-segment processing during Okazaki fragment maturation. Biochemistry. 42, 15045–15052.
Gary R., Kim K., Cornelius H.L., Park M.S., Matsumoto Y. 1999. Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J. Biol. Chem. 274, 4354–4363.
Gomes X.V., Burgers P.M. 2000. Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J. 19, 381138–381121.
Zheng L., Dai H., Qiu J., Huang Q., Shen B. 2007. Disruption of the FEN-1/PCNA interaction results in DNA replication defects, pulmonary hypoplasia, pancytopenia and newborn lethality in mice. Mol. Cell. Biol. 27, 3176–3186.
Parrish J., Li L., Klotz K., Ledwich D., Wang X., Xue D. 2001. Mitochondrial endonuclease G is important for apoptosis in C. elegans. Nature. 412, 90–94.
Parrish J.Z., Xue D. 2003. Functional genomic analysis of apoptotic DNA degradation in C. elegans. Mol. Cell. 11, 987–996.
Murray J.M., Tavassoli M., al-Harithy R., Sheldrick K.S., Lehmann A.R., Carr A.M., Watts F.Z. 1994. Structural and functional conservation of the human homolog of the Schizosaccharomyces pombe rad2 gene, which is required for chromosome segregation and recovery from DNA damage. Mol. Cell. Biol. 14, 4878–4888.
Reagan M.S., Pittenger C., Siede W., Friedberg E.C. 1995. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bacteriol. 177, 364–371.
Vallen E.A., Cross F.R. 1995. Mutations in RAD27 define a potential link between G1 cyclins and DNA replication. Mol. Cell. Biol. 15, 4291–4302.
Otto C.J., Almqvist E., Hayden M.R., Andrew S.E. 2001. The “flap” endonuclease gene FEN1 is excluded as a candidate gene implicated in the CAG repeat expansion underlying Huntington disease. Clin. Genet. 59, 122–127.
Warbrick E., Coates P.J., Hall P.A. 1998. Fen1 expression: A novel marker for cell proliferation. J. Pathol. 186, 319–324.
Kim I., Lee M., Lee I., Shin S., Lee S. 2000. Gene expression of flap endonuclease-1 during cell proliferation and differentiation. Biochim. Biophys. Acta. 1496, 333–340.
Rumbaugh J.A., Henricksen L.A., DeMott M.S., Bambara R.A. 1999. Cleavage of substrates with mismatched nucleotides by flap endonuclease-1. J. Biol. Chem. 274, 14602–14608.
Ayyagari R., Gomes X.V., Gordenin D.A., Burgers P.M. 2003. Okazaki fragment maturation in yeast: 1. Distribution of functions between FEN1 and DNA2. J. Biol. Chem. 278, 1618–1625.
Sommers C.H., Miller E.J., Dujon B., Prakash S., Prakash L. 1995. Conditional lethality of null mutations in RTH1 that encodes the yeast counterpart of a mammalian 5′-to 3′-exonuclease required for lagging strand DNA synthesis in reconstituted systems. J. Biol. Chem. 270, 4193–4196.
Rossi M.L., Bambara R.A. 2006. Reconstituted Okazaki fragment processing indicates two pathways of primer removal. J. Biol. Chem. 281, 26051–26061.
Budd M.E., Choe W., Campbell J.L. 2000. The nuclease activity of the yeast DNA2 protein, which is related to the RecB-like nucleases, is essential in vivo. J. Biol. Chem. 275, 16518–16529.
Lee K.H., Kim D.W., Bae S.H., Kim J.A., Ryu G.H., Kwon Y.N., Kim K.A., Koo H.S., Seo Y.S. 2000. The endonuclease activity of the yeast Dna2 enzyme is essential in vivo. Nucleic Acids Res. 28, 2873–2881.
Kang H.Y., Choi E., Bae S.H., Lee K.H., Gim B.S., Kim H.D., Park C., MacNeill S.A., Seo Y.S. 2000. Genetic analyses of Schizosaccharomyces pombe dna2(+) reveal that dna2 plays an essential role in Okazaki fragment metabolism. Genetics. 155, 1055–1067.
Kao H.I., Campbell J.L., Bambara R.A. 2004. Dna2p helicase/nuclease is a tracking protein, like FEN1, for flap cleavage during Okazaki fragment maturation. J. Biol. Chem. 279, 50840–50849.
Bae S.H., Seo Y.S. 2000. Characterization of the enzymatic properties of the yeast dna2 helicase/endonuclease suggests a new model for Okazaki fragment processing. J. Biol. Chem. 275, 38022–38031.
Rossi M.L., Purohit V., Brandt P.D., Bambara R.A. 2006. Lagging strand replication proteins in genome stability and DNA repair. Chem. Rev. 106, 453–473.
Bennett P. 2000. Demystified … microsatellites. Mol. Pathol. 53, 177–183.
Henricksen L.A., Tom S., Liu Y., Bambara R.A. 2000. Inhibition of flap endonuclease 1 by flap secondary structure and relevance to repeat sequence expansion. J. Biol. Chem. 275, 16 420–16 427.
Liu Y., Zhang H., Veeraraghavan J., Bambara R.A., Freudenreich C.H. 2004. Saccharomyces cerevisiae flap endonuclease 1 uses flap equilibration to maintain triplet repeat stability. Mol. Cell. Biol. 24, 4049–4064.
Singh P., Zheng L., Chavez V., Qiu J., Shen B. 2007. Concerted action of exonuclease and gap-dependent endonuclease activities of FEN-1 contributes to the resolution of triplet repeat sequences (CTG)n-and (GAA)n-derived secondary structures formed during maturation of Okazaki fragments. J. Biol. Chem. 282, 3465–3477.
Barnes C.J., Walf A.F., Shen B., Park M.S., Bambara R.A. 1996. Mechanism of tracking and cleavage of adduct-damaged DNA substrates by the mammalian 5′-to 3′-exonuclease/endonuclease RAD2 homologue 1 or flap endonuclease 1. J. Biol. Chem. 271, 29624–29631.
Yoon J.H., Swiderski P.M., Kaplan B.E., Takao M., Yasui A., Shen B., Pfeifer G.P. 1999. Processing of UV damage in vitro by FEN-1 proteins as part of an alternative DNA excision repair pathway. Biochemistry. 38, 4809–4817.
Aboussekhra A., Wood R.D. 1994. Repair of UV-damaged DNA by mammalian cells and Saccharomyces cerevisiae. Curr. Opin. Genet. Dev. 4, 212–220.
Tseng H.-M., Tomkinson A.E. 2004. Processing and joining of DNA ends coordinated by interactions among Dnl4/Lif1, Pol4, and FEN-1. J. Biol. Chem. 279, 47580–47588.
Schaarer O.D. 2003. Chemistry and biology of DNA repair. Angew. Chem. Int. Ed. 42, 2946–2974.
Wu X., Wilson T.E., Lieber M.R. 1999. A role for FEN-1 in nonhomologous DNA end joining: The order of strand annealing and nucleolytic processing events. Proc. Natl. Acad. Sci. USA. 96, 1303–1308.
Critchlow S.E., Jackson S.P. 1998. DNA end-joining: From yeast to man. Trends Biochem. Sci. 23, 394–398.
Wilson D.M. III, Thompson L.H. 1997. Life without DNA repair. Proc. Natl. Acad. Sci. USA. 94, 12754–12757.
DeMott M.S., Shen B.H., Park M.S., Bambara R.A., Zigman S. 1996. Human RAD2 homolog 1 5′-to 3′-exo/endonuclease can efficiently excise a displaced DNA fragment containing a 5′-terminal abasic lesion by endonuclease activity. J. Biol. Chem. 271, 30068–30076.
Klungland A., Lindahl T. 1997. Second pathway for completion of human DNA base excision-repair: Reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J. 16, 3341–3348.
Frosina G., Fortini P., Rossi O., Carrozzino F., Raspaglio G. 1996. Two pathways for base excision repair in mammalian cells. J. Biol. Chem. 271, 9573–9578.
Kim K., Biade S., Matsumoto Y. 1998. Involvement of flap endonuclease 1 in base excision DNA repair. J. Biol. Chem. 273, 8842–8848.
Stucki M., Pascucci B., Parlanti E., Fortini P., Wilson S.H., Hubscher U., Dogliotti E. 1998. Mammalian base excision repair by DNA polymerases delta and epsilon. Oncogene. 17, 835–843.
Tom S., Ranalli T.A., Podust V.N., Bambara R.A. 2001. Regulatory roles of p21 and apurinic/apyrimidinic endonuclease 1 in base excision repair. J. Biol. Chem. 276, 48781–48789.
Khodyreva S.N., Lavrik O.I. 2005. Photoaffinity labeling technique for studying DNA replication and DNA repair. Curr. Med. Chem. 12, 641–655.
Lavrik O.I., Prasad R., Sobol R.W., Horton J.K., Ackerman E.J., Wilson S.H. 2001. Photoaffinity labeling of mouse fibroblast enzymes by a base excision repair intermediate. J. Biol. Chem. 27, 25541–25548.
Liu Y., Beard W.A., Shock D.D., Prasad R., Hou E.W., Wilson S.H. 2005. DNA polymerase β and flap endonuclease 1 enzymatic specificities sustain DNA synthesis for long patch base excision repair. J. Biol. Chem. 280, 3665–3674.
Lebedeva N.A., Rechkunova N.I., Dezhurov S.V., Khodyreva S.N., Favre A., Blanco L., Lavrik O.I. 2005. Comparison of functional properties of mammalian DNA polymerase λ and DNA polymerase β in reactions of DNA synthesis related to DNA repair. Biochim. Biophys. Acta. 1751, 150–158.
Zheng L., Dai H., Zhou M., Li M., Singh P., Qiu J., Tsark W., Huang Q., Kernstine K., Zhang X., Lin D., Shen B. 2007. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat. Medicine. 13, 812–819.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © Zh.K. Nazarkina, O.I. Lavrik, S.N. Khodyreva, 2008, published in Molekulyarnaya Biologiya, 2008, Vol. 42, No. 3, pp. 405–421.
Rights and permissions
About this article
Cite this article
Nazarkina, Z.K., Lavrik, O.I. & Khodyreva, S.N. Flap endonuclease 1 and its role in eukaryotic DNA metabolism. Mol Biol 42, 357–370 (2008). https://doi.org/10.1134/S0026893308030035
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893308030035