Abstract
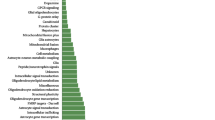
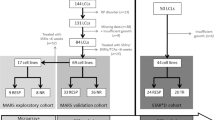
Drug-effect phenotypes in human lymphoblastoid cell lines recently allowed to identify CHL1 (cell adhesion molecule with homology to L1CAM), GAP43 (growth-associated protein 43) and ITGB3 (integrin beta 3) as new candidates for involvement in the antidepressant effect. CHL1 and ITGB3 code for adhesion molecules, while GAP43 codes for a neuron-specific cytosolic protein expressed in neuronal growth cones; all the three gene products are involved in synaptic plasticity. Sixteen polymorphisms in these genes were genotyped in two samples (n=369 and 90) with diagnosis of major depressive episode who were treated with antidepressants in a naturalistic setting. Phenotypes were response, remission and treatment-resistant depression. Logistic regression including appropriate covariates was performed. Genes associated with outcomes were investigated in the Sequenced Treatment Alternatives to Relieve Depression (STAR*D) genome-wide study (n=1861) as both individual genes and through a pathway analysis (Reactome and String databases). Gene-based analysis suggested CHL1 rs4003413, GAP43 rs283393 and rs9860828, ITGB3 rs3809865 as the top candidates due to their replication across the largest original sample and the STAR*D cohort. GAP43 molecular pathway was associated with both response and remission in the STAR*D, with ELAVL4 representing the gene with the highest percentage of single nucleotide polymorphisms (SNPs) associated with outcomes. Other promising genes emerging from the pathway analysis were ITGB1 and NRP1. The present study was the first to analyze cell adhesion genes and their molecular pathways in antidepressant response. Genes and biomarkers involved in neuronal adhesion should be considered by further studies aimed to identify predictors of antidepressant response.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Judd LL, Akiskal HS, Zeller PJ, Paulus M, Leon AC, Maser JD et al. Psychosocial disability during the long-term course of unipolar major depressive disorder. Arch Gen Psychiatry 2000; 57: 375–380.
Franchini L, Serretti A, Gasperini M, Smeraldi E . Familial concordance of fluvoxamine response as a tool for differentiating mood disorder pedigrees. J Psychiatr Res 1998; 32: 255–259.
Tansey KE, Guipponi M, Hu X, Domenici E, Lewis G, Malafosse A et al. Contribution of common genetic variants to antidepressant response. Biol Psychiatry 2013; 73: 679–682.
Fabbri C, Marsano A, Albani D, Chierchia A, Calati R, Drago A et al. PPP3CC gene: a putative modulator of antidepressant response through the B-cell receptor signaling pathway. Pharmacogenomics J 2014; 14: 463–472.
Maness PF, Schachner M . Neural recognition molecules of the immunoglobulin superfamily: signaling transducers of axon guidance and neuronal migration. Nat Neurosci 2007; 10: 19–26.
Morellini F, Lepsveridze E, Kahler B, Dityatev A, Schachner M . Reduced reactivity to novelty, impaired social behavior, and enhanced basal synaptic excitatory activity in perforant path projections to the dentate gyrus in young adult mice deficient in the neural cell adhesion molecule CHL1. Mol Cell Neurosci 2007; 34: 121–136.
Demyanenko GP, Siesser PF, Wright AG, Brennaman LH, Bartsch U, Schachner M et al. L1 and CHL1 cooperate in thalamocortical axon targeting. Cereb Cortex 2011; 21: 401–412.
Demyanenko GP, Halberstadt AI, Rao RS, Maness PF . CHL1 cooperates with PAK1-3 to regulate morphological differentiation of embryonic cortical neurons. Neuroscience 2010; 165: 107–115.
Desarnaud F, Jakovcevski M, Morellini F, Schachner M . Stress downregulates hippocampal expression of the adhesion molecules NCAM and CHL1 in mice by mechanisms independent of DNA methylation of their promoters. Cell Adh Migr 2008; 2: 38–44.
Noor A, Lionel AC, Cohen-Woods S, Moghimi N, Rucker J, Fennell A et al. Copy number variant study of bipolar disorder in Canadian and UK populations implicates synaptic genes. Am J Med Genet B Neuropsychiatr Genet 2014; 165B: 303–313.
Tam GW, van de Lagemaat LN, Redon R, Strathdee KE, Croning MD, Malloy MP et al. Confirmed rare copy number variants implicate novel genes in schizophrenia. Biochem Soc Trans 2010; 38: 445–451.
Chu TT, Liu Y . An integrated genomic analysis of gene-function correlation on schizophrenia susceptibility genes. J Hum Genet 2010; 55: 285–292.
Morag A, Pasmanik-Chor M, Oron-Karni V, Rehavi M, Stingl JC, Gurwitz D . Genome-wide expression profiling of human lymphoblastoid cell lines identifies CHL1 as a putative SSRI antidepressant response biomarker. Pharmacogenomics 2011; 12: 171–184.
Oved K, Morag A, Pasmanik-Chor M, Oron-Karni V, Shomron N, Rehavi M et al. Genome-wide miRNA expression profiling of human lymphoblastoid cell lines identifies tentative SSRI antidepressant response biomarkers. Pharmacogenomics 2012; 13: 1129–1139.
Cingolani LA, Goda Y . Differential involvement of beta3 integrin in pre- and postsynaptic forms of adaptation to chronic activity deprivation. Neuron Glia Biol 2008; 4: 179–187.
Pozo K, Cingolani LA, Bassani S, Laurent F, Passafaro M, Goda Y . Beta3 integrin interacts directly with GluA2 AMPA receptor subunit and regulates AMPA receptor expression in hippocampal neurons. Proc Natl Acad Sci USA 2012; 109: 1323–1328.
Wang KS, Liu X, Arana TB, Thompson N, Weisman H, Devargas C et al. Genetic association analysis of ITGB3 polymorphisms with age at onset of schizophrenia. J Mol Neurosci 2013; 51: 446–453.
Napolioni V, Lombardi F, Sacco R, Curatolo P, Manzi B, Alessandrelli R et al. Family-based association study of ITGB3 in autism spectrum disorder and its endophenotypes. Eur J Hum Genet 2011; 19: 353–359.
Carneiro AM, Cook EH, Murphy DL, Blakely RD . Interactions between integrin alphaIIbbeta3 and the serotonin transporter regulate serotonin transport and platelet aggregation in mice and humans. J Clin Invest 2008; 118: 1544–1552.
Whyte A, Jessen T, Varney S, Carneiro AM . Serotonin transporter and integrin beta 3 genes interact to modulate serotonin uptake in mouse brain. Neurochem Int 2014; 73: 122–126.
Verhaagen J, Oestreicher AB, Grillo M, Khew-Goodall YS, Gispen WH, Margolis FL . Neuroplasticity in the olfactory system: differential effects of central and peripheral lesions of the primary olfactory pathway on the expression of B-50/GAP43 and the olfactory marker protein. J Neurosci Res 1990; 26: 31–44.
Leu B, Koch E, Schmidt JT . GAP43 phosphorylation is critical for growth and branching of retinotectal arbors in zebrafish. Dev Neurobiol 2010; 70: 897–911.
Zaccaria KJ, Lagace DC, Eisch AJ, McCasland JS . Resistance to change and vulnerability to stress: autistic-like features of GAP43-deficient mice. Genes Brain Behav 2010; 9: 985–996.
Fung SJ, Sivagnanasundaram S, Weickert CS . Lack of change in markers of presynaptic terminal abundance alongside subtle reductions in markers of presynaptic terminal plasticity in prefrontal cortex of schizophrenia patients. Biol Psychiatry 2011; 69: 71–79.
Oved K, Morag A, Pasmanik-Chor M, Rehavi M, Shomron N, Gurwitz D . Genome-wide expression profiling of human lymphoblastoid cell lines implicates integrin beta-3 in the mode of action of antidepressants. Transl Psychiatry 2013; 3: e313.
Souery D, Oswald P, Massat I, Bailer U, Bollen J, Demyttenaere K et al. Clinical factors associated with treatment resistance in major depressive disorder: results from a European multicenter study. J Clin Psychiatry 2007; 68: 1062–1070.
Sheehan DV, Lecrubier Y, Sheehan KH, Amorim P, Janavs J, Weiller E et al. The Mini-International Neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry 1998; 59: 22–33.
Fabbri C, Marsano A, Balestri M, De Ronchi D, Serretti A . Clinical features and drug induced side effects in early versus late antidepressant responders. J Psychiatr Res 2013; 47: 1309–1318.
Howland RH . Sequenced Treatment Alternatives to Relieve Depression (STAR*D). Part 1: study design. J Psychosoc Nurs Ment Health Serv 2008; 46: 21–24.
Trivedi MH, Rush AJ, Ibrahim HM, Carmody TJ, Biggs MM, Suppes T et al. The Inventory of Depressive Symptomatology, Clinician Rating (IDS-C) and Self-Report (IDS-SR), and the Quick Inventory of Depressive Symptomatology, Clinician Rating (QIDS-C) and Self-Report (QIDS-SR) in public sector patients with mood disorders: a psychometric evaluation. Psychol Med 2004; 34: 73–82.
CHMP. Note for guidance on clinical investigation of medicinal products in the treatment of depression 2002. Available from http://www.emea.europa.eu/pdfs/human/ewp/051897en.pdf.
Thase ME . The need for clinically relevant research on treatment-resistant depression. J Clin Psychiatry 2001; 62: 221–224.
Fabbri C, Drago A, Serretti A . Early antidepressant efficacy modulation by glutamatergic gene variants in the STAR*D. Eur Neuropsychopharmacol 2012; 23: 612–621.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT . Data quality control in genetic case-control association studies. Nat Protoc 2010; 5: 1564–1573.
Bronicki LM, Jasmin BJ . Emerging complexity of the HuD/ELAVl4 gene; implications for neuronal development, function, and dysfunction. RNA 2013; 19: 1019–1037.
Pechnick RN, Zonis S, Wawrowsky K, Cosgayon R, Farrokhi C, Lacayo L et al. Antidepressants stimulate hippocampal neurogenesis by inhibiting p21 expression in the subgranular zone of the hipppocampus. PLoS One 2011; 6: e27290.
Epp JR, Beasley CL, Galea LA . Increased hippocampal neurogenesis and p21 expression in depression: dependent on antidepressants, sex, age, and antipsychotic exposure. Neuropsychopharmacology 2013; 38: 2297–2306.
Pedram A, Razandi M, Deschenes RJ, Levin ER . DHHC-7 and -21 are palmitoylacyltransferases for sex steroid receptors. Mol Biol Cell 2012; 23: 188–199.
Ooishi Y, Kawato S, Hojo Y, Hatanaka Y, Higo S, Murakami G et al. Modulation of synaptic plasticity in the hippocampus by hippocampus-derived estrogen and androgen. J Steroid Biochem Mol Biol 2012; 131: 37–51.
Parker G, Brotchie H . Gender differences in depression. Int Rev Psychiatry 2010; 22: 429–436.
Chandley MJ, Szebeni A, Szebeni K, Crawford JD, Stockmeier CA, Turecki G et al. Elevated gene expression of glutamate receptors in noradrenergic neurons from the locus coeruleus in major depression. Int J Neuropsychopharmacol 2014: 1–10.
Mazalouskas M, Jessen T, Varney S, Sutcliffe JS, Veenstra-VanderWeele J, Cook EH Jr et al. Integrin β3 Haploinsufficiency Modulates Serotonin Transport and Antidepressant-Sensitive Behavior in Mice. Neuropsychopharmacology 2015, [Epub ahead of print].
Ikeda A, Kato T . Biological predictors of lithium response in bipolar disorder. Psychiatry Clin Neurosci 2003; 57: 243–250.
Rantamaki T, Hendolin P, Kankaanpaa A, Mijatovic J, Piepponen P, Domenici E et al. Pharmacologically diverse antidepressants rapidly activate brain-derived neurotrophic factor receptor TrkB and induce phospholipase-Cgamma signaling pathways in mouse brain. Neuropsychopharmacology 2007; 32: 2152–2162.
Kolodkin AL, Levengood DV, Rowe EG, Tai YT, Giger RJ, Ginty DD . Neuropilin is a semaphorin III receptor. Cell 1997; 90: 753–762.
Pasterkamp RJ, Giger RJ . Semaphorin function in neural plasticity and disease. Curr Opin Neurobiol 2009; 19: 263–274.
Gould TD, Picchini AM, Einat H, Manji HK . Targeting glycogen synthase kinase-3 in the CNS: implications for the development of new treatments for mood disorders. Curr Drug Targets 2006; 7: 1399–1409.
Goswami DB, Jernigan CS, Chandran A, Iyo AH, May WL, Austin MC et al. Gene expression analysis of novel genes in the prefrontal cortex of major depressive disorder subjects. Prog Neuropsychopharmacol Biol Psychiatry 2013; 43: 126–133.
Warner-Schmidt JL, Duman RS . VEGF is an essential mediator of the neurogenic and behavioral actions of antidepressants. Proc Natl Acad Sci USA 2007; 104: 4647–4652.
Warner-Schmidt JL, Duman RS . VEGF as a potential target for therapeutic intervention in depression. Curr Opin Pharmacol 2008; 8: 14–19.
Golden SA, Christoffel DJ, Heshmati M, Hodes GE, Magida J, Davis K et al. Epigenetic regulation of RAC1 induces synaptic remodeling in stress disorders and depression. Nat Med 2013; 19: 337–344.
Acknowledgements
The present study was supported by the grant: Pharmacogenomics of Antidepressant Drug ResponsE (PADRE): tentative drug response biomarkers from human lymphoblastoid cells from ERA-NET NEURON. Data and biomaterials were obtained from the limited access datasets distributed from the NIH-supported ‘Sequenced Treatment Alternatives to Relieve Depression’ (STAR*D). STAR*D focused on non-psychotic major depressive disorder in adults seen in outpatient settings. The primary purpose of this research study was to determine which treatments work best if the first treatment with medication does not produce an acceptable response. The study was supported by NIMH Contract # N01MH90003 to the University of Texas Southwestern Medical Center. The ClinicalTrials.gov identifier is NCT00021528.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
AS is or has been a consultant/speaker for: Abbott, Astra Zeneca, Clinical Data, Boehringer, Bristol-Myers Squibb, Eli Lilly, GlaxoSmithKline, Janssen, Lundbeck, Pfizer, Sanofi and Servier. All other authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website
Supplementary information
PowerPoint slides
Rights and permissions
About this article
Cite this article
Fabbri, C., Crisafulli, C., Gurwitz, D. et al. Neuronal cell adhesion genes and antidepressant response in three independent samples. Pharmacogenomics J 15, 538–548 (2015). https://doi.org/10.1038/tpj.2015.15
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tpj.2015.15
This article is cited by
-
Disease-driven top predator decline affects mesopredator population genomic structure
Nature Ecology & Evolution (2024)
-
Cntnap2-dependent molecular networks in autism spectrum disorder revealed through an integrative multi-omics analysis
Molecular Psychiatry (2023)
-
Body mass index interacts with a genetic-risk score for depression increasing the risk of the disease in high-susceptibility individuals
Translational Psychiatry (2022)
-
The Potential Use of Peripheral Blood Mononuclear Cells as Biomarkers for Treatment Response and Outcome Prediction in Psychiatry: A Systematic Review
Molecular Diagnosis & Therapy (2021)
-
Comparative proteomic analysis of the brain and colon in three rat models of irritable bowel syndrome
Proteome Science (2020)