Abstract
Multiple osteochondromas is an autosomal dominant skeletal disorder characterized by the formation of multiple cartilage-capped tumours. Two causal genes have been identified, EXT1 and EXT2, which account for 65% and 30% of cases, respectively. We have undertaken a mutation analysis of the EXT1 and EXT2 genes in 39 unrelated Spanish patients, most of them with moderate phenotype and looked for genotype-phenotype correlations. We found the mutant allele in 37 patients, 29 in EXT1 and 8 in EXT2. Five of the EXT1 mutations were deletions identified by MLPA. Two cases of mosaicism were documented. We detected a lower number of exostoses in patients with missense mutation versus other kinds of mutations. In conclusion, we found a mutation in EXT1 or in EXT2 in 95% of the Spanish patients. Eighteen of the mutations were novel.
Similar content being viewed by others
Introduction
Multiple osteochondromas (MO, MIM#133700, #133701), also known as multiple hereditary exostoses, is an autosomal dominant skeletal disorder characterized by the formation of multiple cartilage-capped tumours (exostoses or osteochondromas). The prevalence of MO is estimated at 1/50,000 in the Western population1. Osteochondromas are the result of excessive chondrocyte proliferation and bone growth at the juxtaepiphyseal regions of long bones and are the most common benign bone tumour. MO is characterized by an important inter- and intra-familial phenotypic variability, including variations in the number and size of osteochondromas and in the number and location of bones involved. Secondary complications are heterogeneous too and may consist of deformities, functional limitations, compression of nerves and blood vessels, pain caused by pressure on neighbouring tissues and short stature2. The most serious secondary complication is the malignant transformation toward a secondary peripheral chondrosarcoma, occurring in 0.5–5% of patients3.
MO is a genetically heterogeneous disease. In almost 90% of MO patients, germline mutations in EXT1 (MIM #608177)4 or EXT2 (MIM #608210)5,6 are found. EXT1 maps to chromosome 8q24.11–q24.13, comprises 11 exons and spans approximately 350 kb7, while EXT2, located at 11p11–p11.2, consists of 16 exons and spans almost 108 kb8. Both genes act as tumour suppressors. Involvement of other genes has been suggested, since there are families with no mutation identified in either EXT1 or EXT2.
The EXT genes encode ubiquitous glycosyltransferases, catalyzing heparan sulfate (HS) polymerization at HS proteoglycans (HSPGs). These HSPGs play a major role in the diffusion of signalling molecules such as Indian Hedgehog (Ihh), which is an important regulator of chondrocyte proliferation and differentiation in the growth plate9.
Approximately 10% of patients have a de novo mutation10. Loss of the wild-type allele in hereditary cases indicates that inactivation of both EXT alleles is required for osteochondroma formation11, confirming their tumour suppressor action that results in a loss of chondrocyte polarization12. However, the inactivation of both alleles probably occurs only in some of the cells in the cartilaginous cap of osteochondromas12,13.
Several studies have reported on MO causing mutations in different populations. These are being gathered in the Multiple Osteochondromas Mutation Database (MOdb) (http://medgen.ua.ac.be/LOVDv.2.0/), currently listing over 400 and 200 different mutations in EXT1 and EXT2, respectively. Most of the mutations (80%) are nonsense, frame-shift and splice-site mutations, resulting in a premature termination of translation, or involve partial or total deletion of the gene. Although it has been suggested that mutations in EXT1 are associated with a more severe phenotype than mutations in EXT214,15,16, many aspects of the phenotypic variability observed in patients have yet to be understood at the genetic level. As suggested by the Human Variome Project initiative, characterization of causative mutations in familial and sporadic cases in diverse populations is needed for full understanding of Mendelian diseases17.
Here we present the mutational analysis of 39 unrelated Spanish MO patients and the clinical features of most of them. This is the first report of MO mutations in the Spanish population.
Results
Clinical features
The main clinical features of the patients are detailed in Table 1. Most of the patients (77%) had deformities (classes II and III). More than half the cases (51%) had more than 20 exostoses. Mean age of onset was 2 years and clinical evaluation was performed, on average, 12 years later. Only one of the 39 cases developed a malignant transformation.
EXT1 and EXT2 point mutations
On sequencing all exons and flanking regions of the EXT1 and EXT2 genes in the samples of 39 unrelated patients, 31 pathogenic point mutations were identified (Table 2). Twenty-four different mutations were found in the EXT1 gene, while 7 were found in EXT2. One of them (EXT2, c.544C > T) was found in two unrelated patients. Eighteen out of the 31 mutations were novel, two of them missense in EXT1: p.Asp231Val and p.Pro337Arg. Bioinformatic predictions suggested a pathogenic role for both of them [PolyPhen, probably_damaging (1); SIFT, deleterious (0)]. The mutation screening also detected two changes that were interpreted as non-pathogenic (EXT1, c.962+8_962+11delTCTG and EXT2, c.1178G > A), indicated in italics in Table 2. These two changes were found in patients BCN33 and BCN29, respectively, together with additional mutations, as indicated in Figure 1 and Table 2. In the case of BCN33, the patient inherited the EXT1, c.962+8_962+11delTCTG change from his unaffected mother, while the other mutation, EXT2, c.544C > T (p.Arg182*), was putatively inherited from his affected father (from whom a sample was not available). Patient BCN29 inherited the EXT2, c.1178G > A (p.Arg393Gln) change from her unaffected father, while she presented with a de novo, previously described mutation in the EXT1 gene: c.1019G > C (p.Arg340Pro). After sequencing and thorough manual checking of chromatograms, seven patients remained undiagnosed at the molecular level.
EXT1 deletions
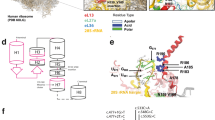
Exon dosage for EXT1 and EXT2 was assessed by MLPA analysis of the seven samples without identified point mutations. Five unrelated patients (BCN06, BCN07, BCN19, BCN20/25 and BCN22) were found to bear different EXT1 deletions (Figure 2 and Table 2). No large deletion was found in the EXT2 gene. The proband from family 20/25 was hemizygous for all EXT1 exons (i.e., he presented with half a dose), while probands BCN06 and BCN07 (the latter not shown in Figure 2) were hemizygous for exons 2 to 11 and patient BCN19 was hemizygous only for exon 8. In contrast, patient BCN22 showed a partial loss of dose for exons 2 to 11, consistent with a mosaic constitution (see Figure 2). Relatives of patient BCN06, including the affected father, were available for MLPA examination. As seen in Figure 2, the father had a partial loss of dose of exons 2 to 11, compared with the unaffected relatives (proband's mother and brother), consistent with a case of mosaicism for the deletion present in his son. For two cases (BCN21 and BCN28) no mutation was found after sequencing and MLPA analyses.
MLPA results for some of the patients bearing deletions of different size in the EXT1 gene.
In the Y axis, 1 and below 0.6 correspond to full dose (two copies) and half dose (one copy) of the corresponding exons, respectively. For the BCN06 family, results for different members of the family are shown. Arrow in BCN19 proband indicates the deletion of exon 8.
Phenotype-genotype correlations
We examined the EXT1 and EXT2 mutations in relation to the various phenotypic aspects shown in Table 1. Gene distribution within the three clinical classes showed a higher proportion of EXT2 mutations in class III than in classes I and II (Figure 3A). No significant differences were found when comparing patients with EXT1 and EXT2 mutations for age of disease onset or number of osteochondromas. In EXT1 patients, we compared the group of 6 individuals bearing missense mutations with 18 patients carrying other type of mutations (nonsense and small or large insertions and deletions) in relation to the number of exostoses. As shown in Figure 3B, a significant difference was found (mean number of exostoses for missense = 14.33; SD = 8.24; mean for other = 26.50; SD = 12.66; p = 0.04). However, no significant difference in the age of onset of the disease was found between these two groups (not shown). Lumbar densitometry was performed in 18 patients: results showed impairment in 6 cases, which harboured non-missense mutations, 4 in EXT1 gene and 2 in EXT2 gene.
Genotype-phenotype correlations.
(A) Distribution of cases with mutations in EXT1 or EXT2 among the three clinical classes. (B) Average number of exostoses among patients bearing missense mutations in EXT1 (n = 6) or other type of mutations (nonsense and small or large insertions and deletions) in the same gene (n = 18).
Patients without identified mutations in EXT1 or EXT2
Details of the two unrelated patients (BCN21 and BCN28) with no identified EXT1 or EXT2 mutations are included in Table 1. Patient BCN21 was a son of an unaffected couple, while BCN28 inherited the disease from his father. Both presented with a number of exostoses and an onset age that are within the range of the rest of the patients.
Discussion
Thirty-nine Spanish unrelated patients were analyzed and the mutant allele was identified in 37 of them. Twenty-nine patients (74%) had mutations in EXT1, while 8 (21%) had mutations in EXT2 (Figure 4A). Only two patients remained undiagnosed at the molecular level. Genotype-phenotype correlations were analysed. Patients bearing EXT1 missense mutations correlated with a lower number of exostoses.
Comparison of mutation frequencies with previous studies.
(A) Proportion of EXT1 mutation cases (dark grey), EXT2 mutation cases (light grey) and cases with no mutation identified (white). (B) Among cases with identified mutations, proportion of large rearrangements (at least one exon) are shown in dark grey, while point mutations and small insertions and deletions are shown in light grey.
The greater number of EXT1 mutations is in agreement with most other studies of different populations (Figure 4A)10,14,18,21,22,23. The proportion of missense mutation (15.4%) agrees with the large study by Pedrini et al.18. All the missense mutations were in EXT1 and 5 out of the 6 clustered in residues 337–341, while the novel missense mutation (p.Asp231Val), lay outside this region. Five probands (14% of the cases) were found to carry partial or whole EXT1 deletions. This proportion is higher than that found by Jennes et al.21 and Signori et al.23 (Figure 4B). All these deletions had been previously described21. However, whether the breakpoints in all cases are the same or not remains to be studied. Only one publication has addressed this issue and found that two cases with deletion of exons 2 to 11 of EXT1 (and two cases with an exon 8 deletion in EXT2) bore different breakpoints21.
We found two cases of mosaicism for the exon 2–11 deletion, which deserve further discussion. In one case, the patient (BCN06) bore the deletion, while the father, from whom he had inherited the disease, was discovered to be a mosaic thanks to the previous finding of the son's mutation. The other case (BCN22) showed results that were consistent with a deletion, although the MLPA values for exons 2 to 11 were above the threshold for hemizygosity (see Figure 2). These cases are similar to those described by Szuhai et al.24. Both mosaic cases (BCN06f and BCN22) had an early disease onset (2 years) and neither was affected less than the average (BCN22 belongs to class IIB). This suggests that mosaic mutations play a role in the pathology: it seems that the deletion of one allele (or of a large part of it) in a relatively small number of cells is enough to trigger the phenotype. However, mosaicism was observed in blood cells, while the actual status in bone remains to be studied.
Two patients bore two changes each, one in EXT1 and one in EXT2. In patient BCN33, the EXT2 nonsense mutation is undoubtedly pathogenic, has been previously described25 and was also found in another patient in our series (BCN18). On the other hand, the pathogenic effect of the donor splice site deletion in intron 1 of EXT1 is dubious. It was described as a pathogenic mutation by Signori et al.23, in an Italian MO patient and was absent in 100 Spanish control chromosomes. However, the splice score was not affected by the change (data not shown). A novel putative donor site generated by the 4-bp deletion has a very low score, making its use unlikely. Moreover, the patient inherited the disease from his father (Figure 1B) and there are several affected relatives on the paternal side. However, the patient inherited the intron 1 4-bp deletion from his healthy mother (and there are no affected relatives on the mother's side). The best explanation with the available data is that the 4-bp deletion in intron 1 is a rare non-pathogenic variant. Patient BCN29 inherited the EXT2 c.1178G > A (p.Arg393Gln) from her unaffected father (Figure 1B). Additionally, she presented with a de novo EXT1 mutation: c.1019G > C (p.Arg340Pro). This mutation has been previously described by several authors and reviewed by Jennes et al.10. That the father had the EXT2 p.Arg393Gln change and is unaffected and that the other mutation was shown to be pathogenic strongly suggest that the former is a non-pathogenic variant. Protein sequence alignments show that Drosophila melanogaster bears a Gln at this position (data not shown). Additionally, the p.Arg393Gln change was found in the Exome Variant Server, NHLBI Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu/EVS/) [June, 2012] with a frequency of 16/13,004 (0.12%).
At the genotype-phenotype level, our data showed that the class with more severe clinical presentations (class III) had a higher proportion of EXT2 mutations than the other classes. However, it must be noted that this may be due to a relatively small number of patients with EXT2 mutations in our study, all of whom bore truncating mutations. When the average number of exostoses in patients with different types of mutations was compared, a significant difference (p < 0.05) was detected between missense mutations and other types of mutation (nonsense and small or large insertions and deletions). Patients with missense mutations had fewer osteochondromas than the rest. A hypothesis to explain this is that some residual activity of the mutant protein may remain only in the case of mutations of amino acid change, which may be enough to produce a small amount of heparan sulfate. This is in agreement with the continuum model suggested by Berger et al.26 which states that protein function can be a continuum related to the level of expression or activity of the tumour suppressor genes rather than to discrete step-by-step changes in gene copy number. One patient out of 39 developed a malignant tumour (2.6%). This figure is within the range of 2–5% described by several authors (see, for example, Bovee3; Pedrini et al.18). There were two other patients in whom no disease-causing mutation could be identified (5.1%). This proportion is lower than that found in most studies, including the large one by Pedrini et al.18. There are different explanations for the undetected mutations, including a possible third locus, the presence of mosaic point mutations gone undetected or the presence of mutations in unexplored regions such as the promoter, recently characterized by Jennes et al.27, or deep intronic.
Overall, our phenotype-genotype correlation results do not agree with those of Pedrini et al.18, which consisted of an association of a mild phenotype with female sex, EXT2 mutations and absence of EXT1/2 mutations. As stated above, the main limitation of our study was the small sample size. Comparisons between groups had not enough statistical power and we could not detect differences, for example, in the association between sex and the severity of the disease. The main challenge, therefore, will be to collect more cases to further investigate the genotype-phenotype correlation in Spanish patients.
To conclude, this is the first genetic study of MO performed in a Spanish cohort. We described a collection of mutations in EXT1 and EXT2 and it was possible to offer diagnosis and genetic counselling to the MO patients and their families. This is an essential first step, in order to gain insight into the bases of the disease and then to develop novel strategies in the search for possible therapies.
Methods
Patients
In this study we investigated patients from 39 Spanish families with MO. Diagnosis, performed at 2 centres (Hospital Sant Joan de Déu, Barcelona, n = 35 families and Hospital Universitario "La Paz", Madrid, n = 4 families), was based on the presence of MO confirmed by physical and/or radiographic examinations. Blood samples were obtained from patients and available relatives for genomic DNA extraction, after informed consent. Samples from patients were obtained in accordance with the Helsinki Declaration, as revised in 2000. The experiments were approved by the Ethics Committee of Hospital Sant Joan de Déu.
Clinical studies
A clinical diagnosis of MO was achieved after obtaining an accurate medical history and physically examining the patient, including the evaluation of all palpable lesions, height, long bone and trunk deformities and functional limitations.
Patients were classified in three clinical classes based on the presence of deformities and functional limitations (I: no deformities and no functional limitations; II: deformities and no functional limitations; and III: deformities and functional limitations), as reported elsewhere18. The subcategories were defined by the number of osteochondromas (A: less than 20; B: more than 20).
Genotyping and mutation analysis
Genomic DNA mutation screening of the EXT1 and EXT2 genes was performed for each patient. New primers were designed to amplify all exons and flanking intronic regions of both the EXT1 and the EXT2 genes, so that all fragments, except those corresponding to exon 1 of EXT1, could be amplified by polymerase chain reaction (PCR) simultaneously. Primers are listed in on-line Supplementary Table S1. Exon 1 of EXT1 and exon 4 of EXT2 were split into several overlapping fragments, to obtain amplification products that did not exceed 650 bp. PCR was performed in a 50 μl reaction volume, containing ~100 ng of genomic DNA, 1× PCR buffer, 1–2 mM MgCl2, 0.2 mM of each dNTP, 0.4 μM of each forward and reverse primer and 0.7 U of GoTag® Flexi polymerase (Promega, Madison, WI). All PCR programs included an initial denaturation of 4 min at 95°C, followed by 35 cycles of 30 sec at 95°C, 30 sec at annealing temperature (Ta) and 10 sec at 72°C. Finally, an extension at 72°C was performed for 5 min. Annealing temperature was 60°C for all primer combinations, with the exception of primers for the amplification of overlapping regions of exon 1 of EXT1. For these primer combinations, Ta was set at 55°C for ex1.1 and 57°C for ex1.2 and ex1.3. After amplification, the PCR products were purified using a PCR purification kit (GE Healthcare) and then sequenced with BigDye 3.1 (Applied Biosystems; life technologies) in the following PCR programme: denaturation 1 min at 96°C, 25 cycles of 10 sec at 96°C, 5 sec at 55°C and 4 min at 60°C. The sequences were analyzed with an ABI PRISM 3730 DNA Analyzer (Applied Biosystems life technologies). The presence of all the mutations detected was confirmed by digestion of each PCR product with the corresponding enzyme. None of the novel mutations were found in 50 control samples. All chromatograms of EXT1 and EXT2 exons of negative cases were manually re-analyzed.
The mutations were given the official HGVS nomenclature (www.hgvs.org). As reference sequences, NM_000127.2 for EXT1 and NM_000401.3 for EXT2 were used.
MLPA
The number of copies of the EXT1 and EXT2 genes present in genomic DNA samples was analyzed by the multiplex ligation-dependent probe amplification (MLPA) technique designed by MRC-Holland. We used the commercial kit #P215-B1 and followed the manufacturer's instructions. PCR products were run on an ABI 3730 DNA Analyzer capillary sequencer (Applied Biosystems, Forster City, CA, USA). Peaks were analyzed by means of the Coffalyser v9.4 software. The proportion of each peak relative to the height of all peaks was calculated for each sample and then compared to proportions for the corresponding peak averaged for a set of at least ten normal DNA samples. Ratios of 1.0 were treated as normal copy number. Ratios below 0.6 were considered as deletions. Each positive result was confirmed in a second independent MLPA reaction.
Assessment of functionality of missense mutations
Disease causing potential of missense mutations was evaluated using two different bioinformatic tools: SIFT and PolyPhen19,20.
Statistical analysis
Normal distribution of the data and variance differences were assessed by the Kolmogorov-Smirnoff and Lavenne tests, respectively. Assessment of mean differences was performed using ANOVA. A p value <0.05 was considered significant. The statistical analysis used the PASW 18.0 program.
References
Schmale, G. A., Conrad, E. U. r. & Raskind, W. H. The natural history of hereditary multiple exostoses. J Bone Joint Surg Am. 76, 986–992 (1994).
Hennekam, R. C. Hereditary multiple exostoses. J Med Genet. 28, 262–266 (1991).
Bovee, J. V. Multiple osteochondromas. Orphanet J Rare Dis. 3, 3 (2008).
Ahn, J., Ludecke, H. J., Lindow, S., Horton, W. A., Lee, B. & Wagner, M. J. et al. Cloning of the putative tumour suppressor gene for hereditary multiple exostoses (EXT1). Nat Genet. 11, 137–143 (1995).
Stickens, D., Clines, G., Burbee, D., Ramos, P., Thomas, S. & Hogue, D. et al. The EXT2 multiple exostoses gene defines a family of putative tumour suppressor genes. Nat Genet. 14, 25–32 (1996).
Wuyts, W., Van Hul, W., Wauters, J., Nemtsova, M., Reyniers, E. & Van Hul, E. V. et al. Positional cloning of a gene involved in hereditary multiple exostoses. Hum Mol Genet. 5, 1547–1557 (1996).
Ludecke, H. J., Ahn, J., Lin, X., Hill, A., Wagner, M. J. & Schomburg, L. et al. Genomic organization and promoter structure of the human EXT1 gene. Genomics. 40, 351–354 (1997).
Clines, G. A., Ashley, J. A., Shah, S., Lovett, M. The structure of the human multiple exostoses 2 gene and characterization of homologs in mouse and Caenorhabditis elegans. Genome Res. 7, 359–367 (1997).
Bellaiche, Y., The, I. & Perrimon, N. Tout-velu is a Drosophila homologue of the putative tumour suppressor EXT-1 and is needed for Hh diffusion. Nature. 394, 85–88 (1998).
Jennes, I., Pedrini, E., Zuntini, M., Mordenti, M., Balkassmi, S. & Asteggiano, C. G. et al. Multiple osteochondromas: mutation update and description of the multiple osteochondromas mutation database (MOdb). Hum Mutat. 30, 1620–1627 (2009).
Bovee, J. V., Cleton-Jansen, A. M., Wuyts, W., Caethoven, G., Taminiau, A. H. & Bakker, E. et al. EXT-mutation analysis and loss of heterozygosity in sporadic and hereditary osteochondromas and secondary chondrosarcomas. Am J Hum Genet. 65, 689–698 (1999).
de Andrea, C. E., Wiweger, M., Prins, F., Bovee, J. V., Romeo, S. & Hogendoorn, P. C. Primary cilia organization reflects polarity in the growth plate and implies loss of polarity and mosaicism in osteochondroma. Lab Invest. 90, 1091–1101 (2010).
Jones, K. B., Piombo, V., Searby, C., Kurriger, G., Yang, B. & Grabellus, F. et al. A mouse model of osteochondromagenesis from clonal inactivation of Ext1 in chondrocytes. Proc Natl Acad Sci U S A. 107, 2054–2059 (2010).
Francannet, C., Cohen-Tanugi, A., Le Merrer, M., Munnich, A., Bonaventure, J. & Legeai-Mallet, L. Genotype-phenotype correlation in hereditary multiple exostoses. J Med Genet. 38, 430–434 (2001).
Porter, D. E., Lonie, L., Fraser, M., Dobson-Stone, C., Porter, J. R. & Monaco, A. P. et al. Severity of disease and risk of malignant change in hereditary multiple exostoses. A genotype-phenotype study. J Bone Joint Surg Br. 86, 1041–1046 (2004).
Alvarez, C. M., De Vera, M. A., Heslip, T. R. & Casey, B. Evaluation of the anatomic burden of patients with hereditary multiple exostoses. Clin Orthop Relat Res. 462, 73–79 (2007).
Kaput, J., Cotton, R. G., Hardman, L., Watson, M., Al Aqeel, A. I. & Al-Aama, J. Y. et al. Planning the human variome project: the Spain report. Hum Mutat. 30, 496–510 (2009).
Pedrini, E., Jennes, I., Tremosini, M., Milanesi, A., Mordenti, M. & Parra, A. et al. Genotype-phenotype correlation study in 529 patients with multiple hereditary exostoses: identification of "protective" and "risk" factors. J Bone Joint Surg Am. 93, 2294–2302 (2011).
Ng, P. C. & Henikoff, S. Predicting deleterious amino acid substitutions. Genome Res. 11, 863–874 (2001).
Adzhubei, I. A., Schmidt, S., Peshkin, L., Ramensky, V. E., Gerasimova, A. & Bork, P. et al. A method and server for predicting damaging missense mutations. Nat Methods. 7, 248–249 (2010).
Jennes, I., Entius, M. M., Van Hul, E., Parra, A., Sangiorgi, L. & Wuyts, W. Mutation screening of EXT1 and EXT2 by denaturing high-performance liquid chromatography, direct sequencing analysis, fluorescence in situ hybridization and a new multiplex ligation-dependent probe amplification probe set in patients with multiple osteochondromas. J Mol Diagn. 10, 85–92 (2008).
Reijnders, C. M., Waaijer, C. J., Hamilton, A., Buddingh, E. P., Dijkstra, S. P., Ham, J. et al. No haploinsufficiency but loss of heterozygosity for EXT in multiple osteochondromas. Am J Pathol. 177, 1946–1957 (2010).
Signori, E., Massi, E., Matera, M. G., Poscente, M., Gravina, C. & Falcone, G. et al. A combined analytical approach reveals novel EXT1/2 gene mutations in a large cohort of Italian multiple osteochondromas patients. Genes Chromosomes Cancer. 46, 470–477 (2007).
Szuhai, K., Jennes, I., de Jong, D., Bovee, J. V., Wiweger, M. & Wuyts, W. et al. Tiling resolution array-CGH shows that somatic mosaic deletion of the EXT gene is causative in EXT gene mutation negative multiple osteochondromas patients. Hum Mutat. 32, E2036–2049 (2011).
Dobson-Stone, C., Cox, R. D., Lonie, L., Southam, L., Fraser, M. & Wise, C. et al. Comparison of fluorescent single-strand conformation polymorphism analysis and denaturing high-performance liquid chromatography for detection of EXT1 and EXT2 mutations in hereditary multiple exostoses. Eur J Hum Genet. 8, 24–32 (2000).
Berger, A. H., Knudson, A. G. & Pandolfi, P. P. A continuum model for tumour suppression. Nature. 476, 163–169 (2011).
Jennes, I., Zuntini, M., Mees, K., Palagani, A., Pedrini, E. & De Cock, G. et al. Identification and functional characterization of the human EXT1 promoter region. Gene. 492, 148–159 (2012).
Philippe, C., Porter, D. E., Emerton, M. E., Wells, D. E., Simpson, A. H. & Monaco, A. P. Mutation screening of the EXT1 and EXT2 genes in patients with hereditary multiple exostoses. Am J Hum Genet. 61, 520–528 (1997).
Raskind, W. H., Conrad, E. U., 3rd, Matsushita, M., Wijsman, E. M., Wells, D. E. & Chapman N. et al. Evaluation of locus heterogeneity and EXT1 mutations in 34 families with hereditary multiple exostoses. Hum Mutat. 11, 231–239 (1998).
Seki, H., Kubota, T., Ikegawa, S., Haga, N., Fujioka, F. & Ohzeki, S. et al. Mutation frequencies of EXT1 and EXT2 in 43 Japanese families with hereditary multiple exostoses. Am J Med Genet. 99, 59–62 (2001).
Heinritz, W., Huffmeier, U., Strenge, S., Miterski, B., Zweier, C. & Leinung, S. et al. New Mutations of EXT1 and EXT2 Genes in German Patients with Multiple Osteochondromas. Ann Hum Genet. (2009).
Lonie, L., Porter, D. E., Fraser, M., Cole, T., Wise, C. & Yates, L. et al. Determination of the mutation spectrum of the EXT1/EXT2 genes in British Caucasian patients with multiple osteochondromas and exclusion of six candidate genes in EXT negative cases. Hum Mutat. 27, 1160 (2006).
Acknowledgements
We thank the patients and their families for their enthusiastic participation. The authors are also grateful for the support from the Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER), which is an initiative of the ISCIII. This study was funded in part by grants from the Spanish Ministry of Science and Innovation (SAF2010-15707, SAF2011-25431 and PIB2010AR-00473) and from the Generalitat de Catalunya (2009SGR 971). PS was supported by a grant from the Spanish Ministry of Science and Innovation (FPI).
Author information
Authors and Affiliations
Contributions
Study design: R.A., J.An., M.A.V., C.G.A., S.B., D.G. Collection of data and samples: P.S., A.S., L.M., J.Ar., F.T., J.N. Performance of experiments: P.S., R.U., A.D., J.N., P.L. Data interpretation and analysis: P.S., R.A., P.L., S.B., D.G. Draft composition: P.S., R.A., D.G., S.B. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Table S1
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Sarrión, P., Sangorrin, A., Urreizti, R. et al. Mutations in the EXT1 and EXT2 genes in Spanish patients with multiple osteochondromas. Sci Rep 3, 1346 (2013). https://doi.org/10.1038/srep01346
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep01346
This article is cited by
-
Mutation spectrum of EXT1 and EXT2 in the Saudi patients with hereditary multiple exostoses
Orphanet Journal of Rare Diseases (2021)
-
An analysis of osteoporosis in patients with hereditary multiple exostoses
Osteoporosis International (2020)
-
Large-scale mutational analysis in the EXT1 and EXT2 genes for Japanese patients with multiple osteochondromas
BMC Genetics (2016)
-
Integrated analysis of gene expression and genomic aberration data in osteosarcoma (OS)
Cancer Gene Therapy (2015)
-
Genetic screening of EXT1 and EXT2 in Cypriot families with hereditary multiple osteochondromas
Journal of Genetics (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.