Key Points
-
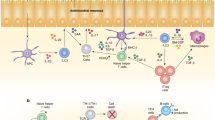
Three distinct groups of innate lymphoid cells (ILCs) have been categorized — these have unique and overlapping functions in immunity.
-
Several committed ILC-restricted progenitor subsets have been discovered, and the transcriptional programmes that drive their generation have also been deciphered.
-
A complex repertoire of transcription factors has been shown to control ILC diversification and maturation.
-
ILC subsets within tissues are maintained and tuned through a set of environmental signals that balance cellular homeostasis and functional plasticity.
Abstract
Innate lymphoid cells (ILCs) are a recently described family of lymphoid effector cells that have important roles in immune defence, inflammation and tissue remodelling. It has been proposed that ILCs represent 'innate' homologues of differentiated effector T cells, and they have been categorized into three groups — namely, ILC1s, ILC2s and ILC3s — on the basis of their expression of cytokines and transcription factors that are typically associated with T helper 1 (TH1)-, TH2- and TH17-type immune responses, respectively. Indeed, remarkable similarity is seen between the specific transcription factors required for the development and diversification of different ILC groups and those that drive effector T cell differentiation. The recent identification of dedicated ILC precursors has provided a view of the mechanisms that control this first essential stage of ILC development. Here, we discuss the transcriptional mechanisms that regulate ILC development and diversification into distinct effector subsets with key roles in immunity and tissue homeostasis. We further caution against the current distinction between 'helper' versus 'killer' subsets in the evolving area of ILC nomenclature.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Spits, H. & Di Santo, J. P. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat. Immunol. 12, 21–27 (2011).
Spits, H. et al. Innate lymphoid cells — a proposal for uniform nomenclature. Nat. Rev. Immunol. 13, 145–149 (2013). This concise review describes and defines the consensus nomenclature for the ILC family.
Di Santo, J. P. Natural killer cell developmental pathways: a question of balance. Annu. Rev. Immunol. 24, 257–286 (2006).
Shi, F. D., Ljunggren, H. G., La Cava, A. & Van Kaer, L. Organ-specific features of natural killer cells. Nat. Rev. Immunol. 11, 658–671 (2011).
Takeda, K. et al. TRAIL identifies immature natural killer cells in newborn mice and adult mouse liver. Blood 105, 2082–2089 (2005).
Tay, C. H. & Welsh, R. M. Distinct organ-dependent mechanisms for the control of murine cytomegalovirus infection by natural killer cells. J. Virol. 71, 267–275 (1997).
Cooper, M. A., Fehniger, T. A. & Caligiuri, M. A. The biology of human natural killer-cell subsets. Trends Immunol. 22, 633–640 (2001).
Fuchs, A. et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity 38, 769–781 (2013).
Klose, C. S. et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell 157, 340–356 (2014).
Sojka, D. K. et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife 3, e01659 (2014).
Daussy, C. et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J. Exp. Med. 211, 563–577 (2014).
Marquardt, N. et al. Cutting edge: identification and characterization of human intrahepatic CD49a+ NK cells. J. Immunol. 194, 2467–2472 (2015).
Bernink, J. H. et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat. Immunol. 14, 221–229 (2013).
Moro, K. et al. Innate production of TH2 cytokines by adipose tissue-associated c-Kit+Sca-1+ lymphoid cells. Nature 463, 540–544 (2010).
Neill, D. R. et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature 464, 1367–1370 (2010).
Price, A. E. et al. Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc. Natl Acad. Sci. USA 107, 11489–11494 (2010).
Halim, T. Y., Krauss, R. H., Sun, A. C. & Takei, F. Lung natural helper cells are a critical source of Th2 cell-type cytokines in protease allergen-induced airway inflammation. Immunity 36, 451–463 (2012).
Ikutani, M. et al. Identification of innate IL-5-producing cells and their role in lung eosinophil regulation and antitumor immunity. J. Immunol. 188, 703–713 (2012).
Fallon, P. G. et al. Identification of an interleukin (IL)-25-dependent cell population that provides IL-4, IL-5, and IL-13 at the onset of helminth expulsion. J. Exp. Med. 203, 1105–1116 (2006).
Bartemes, K. R. et al. IL-33-responsive lineage −CD25+CD44hi lymphoid cells mediate innate type 2 immunity and allergic inflammation in the lungs. J. Immunol. 188, 1503–1513 (2012).
Chang, Y. J. et al. Innate lymphoid cells mediate influenza-induced airway hyper-reactivity independently of adaptive immunity. Nat. Immunol. 12, 631–638 (2011).
Monticelli, L. A. et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat. Immunol. 12, 1045–1054 (2011).
Molofsky, A. B. et al. Innate lymphoid type 2 cells sustain visceral adipose tissue eosinophils and alternatively activated macrophages. J. Exp. Med. 210, 535–549 (2013).
Klein Wolterink, R. G. et al. Essential, dose-dependent role for the transcription factor Gata3 in the development of IL-5+ and IL-13+ type 2 innate lymphoid cells. Proc. Natl Acad. Sci. USA 110, 10240–10245 (2013).
Roediger, B. et al. Cutaneous immunosurveillance and regulation of inflammation by group 2 innate lymphoid cells. Nat. Immunol. 14, 564–573 (2013).
Gold, M. J. et al. Group 2 innate lymphoid cells facilitate sensitization to local, but not systemic, TH2-inducing allergen exposures. J. Allergy Clin. Immunol. 133, 1142–1148 (2014).
Wilhelm, C. et al. An IL-9 fate reporter demonstrates the induction of an innate IL-9 response in lung inflammation. Nat. Immunol. 12, 1071–1077 (2011).
Oliphant, C. J. et al. MHCII-mediated dialog between group 2 innate lymphoid cells and CD4+ T cells potentiates type 2 immunity and promotes parasitic helminth expulsion. Immunity 41, 283–295 (2014).
Halim, T. Y. F. et al. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity 40, 425–435 (2014).
Barlow, J. L. et al. Innate IL-13-producing nuocytes arise during allergic lung inflammation and contribute to airways hyperreactivity. J. Allergy Clin. Immunol. 129, 191–194 (2012).
Salimi, M. et al. A role for IL-25 and IL-33-driven type-2 innate lymphoid cells in atopic dermatitis. J. Exp. Med. 210, 2939–2950 (2013).
Hams, E. et al. IL-25 and type 2 innate lymphoid cells induce pulmonary fibrosis. Proc. Natl Acad. Sci. USA 111, 367–372 (2014).
McHedlidze, T. et al. Interleukin-33-dependent innate lymphoid cells mediate hepatic fibrosis. Immunity 39, 357–371 (2013).
Furusawa, J. et al. Critical role of p38 and GATA3 in natural helper cell function. J. Immunol. 191, 1818–1826 (2013).
Hoyler, T. et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity 37, 634–648 (2012).
Yagi, R. et al. The transcription factor GATA3 is critical for the development of all IL-7Rα-expressing innate lymphoid cells. Immunity 40, 378–388 (2014).
Mjosberg, J. M. et al. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat. Immunol. 12, 1055–1062 (2011).
Liang, H. E. et al. Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nat. Immunol. 13, 58–66 (2012).
Bernink, J. H., Germar, K. & Spits, H. The role of ILC2 in pathology of type 2 inflammatory diseases. Curr. Opin. Immunol. 31, 115–120 (2014).
Brestoff, J. R. et al. Group 2 innate lymphoid cells promote beiging of white adipose tissue and limit obesity. Nature 519, 242–246 (2014).
Lee, M.-W. et al. Activated type 2 innate lymphoid cells regulate beige fat biogenesis. Cell 160, 74–87 (2014).
Walker, J. A., Barlow, J. L. & McKenzie, A. N. Innate lymphoid cells — how did we miss them? Nat. Rev. Immunol. 13, 75–87 (2013).
Van de Pavert, S. A. & Mebius, R. E. New insights into the development of lymphoid tissues. Nat. Rev. Immunol. 10, 664–674 (2010).
Kanamori, Y. et al. Identification of novel lymphoid tissues in murine intestinal mucosa where clusters of c-kit+ IL-7R+ Thy1+ lympho-hemopoietic progenitors develop. J. Exp. Med. 184, 1449–1459 (1996).
Bouskra, D. et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456, 507–510 (2008).
Leavy, O. Mucosal immunology: Multifunctional gut IgA+ plasma cells. Nat. Rev. Immunol. 12, 75 (2012).
Sawa, S. et al. Lineage relationship analysis of RORγt+ innate lymphoid cells. Science 330, 665–669 (2010).
Takatori, H. et al. Lymphoid tissue inducer-like cells are an innate source of IL-17 and IL-22. J. Exp. Med. 206, 35–41 (2009).
Eberl, G. et al. An essential function for the nuclear receptor RORγt in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 5, 64–73 (2004).
Cherrier, M., Sawa, S. & Eberl, G. Notch, Id2, and RORγt sequentially orchestrate the fetal development of lymphoid tissue inducer cells. J. Exp. Med. 209, 729–740 (2012).
Satoh-Takayama, N. et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity 29, 958–970 (2008).
Sanos, S. L. et al. RORγt and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat. Immunol. 10, 83–91 (2009).
Luci, C. et al. Influence of the transcription factor RORγt on the development of NKp46+ cell populations in gut and skin. Nat. Immunol. 10, 75–82 (2009).
Buonocore, S. et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature 464, 1371–1375 (2010).
Klose, C. S. et al. A T-bet gradient controls the fate and function of CCR6−RORγ+ innate lymphoid cells. Nature 494, 261–265 (2013).
Vonarbourg, C. et al. Regulated expression of nuclear receptor RORγt confers distinct functional fates to NK cell receptor-expressing RORγ+ innate lymphocytes. Immunity 33, 736–751 (2010).
Hepworth, M. R. et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature 498, 113–117 (2013).
Neurath, M. F. Cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 14, 329–342 (2014).
Cupedo, T. et al. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORc+ CD127+ natural killer-like cells. Nat. Immunol. 10, 66–74 (2009).
Sonnenberg, G. F. et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science 336, 1321–1325 (2012).
Hoorweg, K. et al. Functional differences between human NKp44− and NKp44+ RORC+ innate lymphoid cells. Front. Immunol. 3, 72 (2012).
Cella, M. et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature 457, 722–725 (2009).
Geremia, A. et al. IL-23-responsive innate lymphoid cells are increased in inflammatory bowel disease. J. Exp. Med. 208, 1127–1133 (2011).
Ng, S. Y.-M., Yoshida, T., Zhang, J. & Georgopoulos, K. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity 30, 493–507 (2009).
Rothenberg, E. V. Transcriptional control of early T and B cell developmental choices. Annu. Rev. Immunol. 32, 283–321 (2014).
Yoshida, H. et al. Expression of α4β7 integrin defines a distinct pathway of lymphoid progenitors committed to T cells, fetal intestinal lymphotoxin producer, NK, and dendritic cells. J. Immunol. 167, 2511–2521 (2001).
Possot, C. et al. Notch signaling is necessary for adult, but not fetal, development of RORγt+ innate lymphoid cells. Nat. Immunol. 12, 949–958 (2011).
Constantinides, M. G., McDonald, B. D., Verhoef, P. A. & Bendelac, A. A committed precursor to innate lymphoid cells. Nature 508, 397–401 (2014). References 9 and 68 are the first reports describing common progenitor cells of ILCs.
Yu, X. et al. The basic leucine zipper transcription factor NFIL3 directs the development of a common innate lymphoid cell precursor. eLife 3, e04406 (2014).
Rosmaraki, E. E. et al. Identification of committed NK cell progenitors in adult murine bone marrow. Eur. J. Immunol. 31, 1900–1909 (2001).
Carotta, S., Pang, S. H. M., Nutt, S. L. & Belz, G. T. Identification of the earliest NK-cell precursor in the mouse BM. Blood 117, 5449–5452 (2011).
Fathman, J. W. et al. Identification of the earliest natural killer cell-committed progenitor in murine bone marrow. Blood 118, 5439–5447 (2011).
Verykokakis, M., Zook, E. C. & Kee, B. L. ID'ing innate and innate-like lymphoid cells. Immunol. Rev. 261, 177–197 (2014).
Boos, M. D., Yokota, Y., Eberl, G. & Kee, B. L. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J. Exp. Med. 204, 1119–1130 (2007).
Yokota, Y. et al. Development of peripheral lymphoid organs and natural killer cells depends on the helix–loop–helix inhibitor Id2. Nature 397, 702–706 (1999).
Ikawa, T., Fujimoto, S., Kawamoto, H., Katsura, Y. & Yokota, Y. Commitment to natural killer cells requires the helix-loop-helix inhibitor Id2. Proc. Natl Acad. Sci. USA 98, 5164–5169 (2001).
Xu, W. et al. E2A transcription factors limit expression of Gata3 to facilitate T lymphocyte lineage commitment. Blood 121, 1534–1542 (2013).
Kwon, K. et al. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity 28, 751–762 (2008).
Kee, B. L. & Murre, C. Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix-loop-helix transcription factor E12. J. Exp. Med. 188, 699–713 (1998).
Heemskerk, M. H. et al. Inhibition of T cell and promotion of natural killer cell development by the dominant negative helix loop helix factor Id3. J. Exp. Med. 186, 1597–1602 (1997).
Nakahiro, T., Kurooka, H., Mori, K., Sano, K. & Yokota, Y. Identification of BMP-responsive elements in the mouse Id2 gene. Biochem. Biophys. Res. Commun. 399, 416–421 (2010).
Tong, X. et al. Transcriptional repressor E4-binding protein 4 (E4BP4) regulates metabolic hormone fibroblast growth factor 21 (FGF21) during circadian cycles and feeding. J. Biol. Chem. 285, 36401–36409 (2010).
Yamajuku, D. et al. Cellular DBP and E4BP4 proteins are critical for determining the period length of the circadian oscillator. FEBS Lett. 585, 2217–2222 (2011).
Male, V., Nisoli, I., Gascoyne, D. M. & Brady, H. J. M. E4BP4: an unexpected player in the immune response. Trends Immunol. 33, 98–102 (2012).
Gascoyne, D. M. et al. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat. Immunol. 10, 1118–1124 (2009).
Kamizono, S. et al. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J. Exp. Med. 206, 2977–2986 (2009).
Geiger, T. L. et al. Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. J. Exp. Med. 211, 1723–1731 (2014).
Seillet, C. et al. Nfil3 is required for the development of all innate lymphoid cell subsets. J. Exp. Med. 211, 1733–1740 (2014).
Xu, W. et al. NFIL3 orchestrates the emergence of common helper innate lymphoid cell precursors. Cell Rep. 10, 2043–2054 (2015). References 69 and 87–89 report the essential role of transcription factor NFIL3 in ILC development.
Ikushima, S. et al. Pivotal role for the NFIL3/E4BP4 transcription factor in interleukin 3-mediated survival of pro-B lymphocytes. Proc. Natl Acad. Sci. USA 94, 2609–2614 (1997).
Seillet, C. et al. Differential requirement for Nfil3 during NK cell development. J. Immunol. 192, 2667–2676 (2014).
Male, V. et al. The transcription factor E4bp4/Nfil3 controls commitment to the NK lineage and directly regulates Eomes and Id2 expression. J. Exp. Med. 211, 635–642 (2014).
Tindemans, I., Serafini, N., Di Santo, J. P. & Hendriks, R. W. GATA-3 function in innate and adaptive immunity. Immunity 41, 191–206 (2014).
Vosshenrich, C. A. et al. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat. Immunol. 7, 1217–1224 (2006).
Serafini, N. et al. Gata3 drives development of RORγt+ group 3 innate lymphoid cells. J. Exp. Med. 211, 199–208 (2014). References 36 and 95 characterized a role for the transcription factor GATA3 in ILC3 differentiation.
Diefenbach, A., Colonna, M. & Koyasu, S. Development, differentiation, and diversity of innate lymphoid cells. Immunity 41, 354–365 (2014).
Samson, S. I. et al. GATA-3 promotes maturation, IFN-γ production, and liver-specific homing of NK cells. Immunity 19, 701–711 (2003).
Constantinides, M. G. & Bendelac, A. Transcriptional regulation of the NKT cell lineage. Curr. Opin. Immunol. 25, 161–167 (2013).
Murphy, K. M. & Reiner, S. L. The lineage decisions of helper T cells. Nat. Rev. Immunol. 2, 933–944 (2002).
Colucci, F. et al. Natural cytotoxicity uncoupled from the Syk and ZAP-70 intracellular kinases. Nat. Immunol. 3, 288–294 (2002).
Magram, J. et al. IL-12-deficient mice are defective in IFN γ production and type 1 cytokine responses. Immunity 4, 471–481 (1996).
Aliahmad, P., de la Torre, B. & Kaye, J. Shared dependence on the DNA-binding factor TOX for the development of lymphoid tissue-inducer cell and NK cell lineages. Nat. Immunol. 11, 945–952 (2010).
Gordon, S. M. et al. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity 36, 55–67 (2012).
Barton, K. et al. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity 9, 555–563 (1998).
Djuretic, I. M. et al. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat. Immunol. 8, 145–153 (2007).
Vahedi, G. et al. Helper T-cell identity and evolution of differential transcriptomes and epigenomes. Immunol. Rev. 252, 24–40 (2013).
Aliahmad, P. & Kaye, J. Development of all CD4 T lineages requires nuclear factor TOX. J. Exp. Med. 205, 245–256 (2008).
Wilkinson, B. et al. TOX: an HMG box protein implicated in the regulation of thymocyte selection. Nat. Immunol. 3, 272–280 (2002).
Sharrocks, A. D. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2, 827–837 (2001).
Muthusamy, N., Barton, K. & Leiden, J. M. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature 377, 639–642 (1995).
Bories, J. C. et al. Increased T-cell apoptosis and terminal B-cell differentiation induced by inactivation of the Ets-1 proto-oncogene. Nature 377, 635–638 (1995).
Ramirez, K. et al. Gene deregulation and chronic activation in natural killer cells deficient in the transcription factor ETS1. Immunity 36, 921–932 (2012).
Wong, S. H. et al. Transcription factor RORα is critical for nuocyte development. Nat. Immunol. 13, 229–236 (2012).
Spooner, C. J. et al. Specification of type 2 innate lymphocytes by the transcriptional determinant Gfi1. Nat. Immunol. 14, 1229–1236 (2013).
Yang, Q. et al. T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity 38, 694–704 (2013).
Huang, Y. et al. IL-25-responsive, lineage-negative KLRG1hi cells are multipotential 'inflammatory' type 2 innate lymphoid cells. Nat. Immunol. 16, 161–169 (2015).
Koyasu, S. Inflammatory ILC2 cells: disguising themselves as progenitors? Nat. Immunol. 16, 133–134 (2015).
Mielke, L. A. et al. TCF-1 controls ILC2 and NKp46+RORγt+ innate lymphocyte differentiation and protection in intestinal inflammation. J. Immunol. 191, 4383–4391 (2013).
Hamilton, B. A. et al. Disruption of the nuclear hormone receptor RORα in staggerer mice. Nature 379, 736–739 (1996).
Halim, T. Y. et al. Retinoic-acid-receptor-related orphan nuclear receptor α is required for natural helper cell development and allergic inflammation. Immunity 37, 463–474 (2012).
Robinette, M. L. et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 16, 306–317 (2015). This paper represents a transcriptional resource for diverse ILC subsets.
Montaldo, E. et al. Human RORγt+CD34+ cells are lineage-specified progenitors of group 3 RORγt+ innate lymphoid cells. Immunity 41, 988–1000 (2014).
Lee, J. S. et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat. Immunol. 13, 144–151 (2012).
Kiss, E. A. et al. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science 334, 1561–1565 (2011).
Qiu, J. et al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity 36, 92–104 (2012).
Van de Pavert, S. A. et al. Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature 508, 123–127 (2014).
Spencer, S. P. et al. Adaptation of innate lymphoid cells to a micronutrient deficiency promotes type 2 barrier immunity. Science 343, 432–437 (2014).
Rankin, L. C. et al. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat. Immunol. 14, 389–395 (2013).
Sciume, G. et al. Distinct requirements for T-bet in gut innate lymphoid cells. J. Exp. Med. 209, 2331–2338 (2012).
Yoshida, H. et al. IL-7 receptor α+ CD3− cells in the embryonic intestine induces the organizing center of Peyer's patches. Int. Immunol. 11, 643–655 (1999).
Bando, J. K., Liang, H.-E. & Locksley, R. M. Identification and distribution of developing innate lymphoid cells in the fetal mouse intestine. Nat. Immunol. 16, 153–160 (2015). This paper describes the identification of ARG1-expressing fetal ILCPs and their progeny.
Vosshenrich, C. A. J. et al. Roles for common cytokine receptor γ-chain-dependent cytokines in the generation, differentiation, and maturation of NK cell precursors and peripheral NK cells in vivo. J. Immunol. 174, 1213–1221 (2005).
Freud, A. G. et al. A human CD34+ subset resides in lymph nodes and differentiates into CD56bright natural killer cells. Immunity 22, 295–304 (2005).
Mackley, E. C. et al. CCR7-dependent trafficking of RORγ+ ILCs creates a unique microenvironment within mucosal draining lymph nodes. Nat. Commun. 6, 5862–5869 (2015).
Puzanov, I. J., Bennett, M. & Kumar, V. IL-15 can substitute for the marrow microenvironment in the differentiation of natural killer cells. J. Immunol. 157, 4282–4285 (1996).
Kennedy, M. K. et al. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J. Exp. Med. 191, 771–780 (2000).
Satoh-Takayama, N. et al. IL-7 and IL-15 independently program the differentiation of intestinal CD3−NKp46+ cell subsets from Id2-dependent precursors. J. Exp. Med. 207, 273–280 (2010).
Huntington, N. D. et al. IL-15 trans-presentation promotes human NK cell development and differentiation in vivo. J. Exp. Med. 206, 25–34 (2009).
Schmutz, S. et al. Cutting edge: IL-7 regulates the peripheral pool of adult ROR γ+ lymphoid tissue inducer cells. J. Immunol. 183, 2217–2221 (2009).
Merzoug, L. B. et al. Conditional ablation of NKp46+ cells using a novel Ncr1greenCre mouse strain: NK cells are essential for protection against pulmonary B16 metastases. Eur. J. Immunol. 44, 3380–3391 (2014).
Ranson, T. et al. IL-15 is an essential mediator of peripheral NK-cell homeostasis. Blood 101, 4887–4893 (2003).
Lodolce, J. P. et al. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity 9, 669–676 (1998).
Liou, Y.-H. et al. Adipocyte IL-15 regulates local and systemic NK cell development. J. Immunol. 193, 1747–1758 (2014).
Kim, G. Y., Hong, C. & Park, J.-H. Seeing is believing: illuminating the source of in vivo interleukin-7. Immune Netw. 11, 1–10 (2011).
Leavy, O. Natural killer cells: adaptive control of NK cells. Nat. Rev. Immunol. 13, 394 (2013).
Sun, J. C., Beilke, J. N. & Lanier, L. L. Adaptive immune features of natural killer cells. Nature 457, 557–561 (2009).
Kanno, Y., Vahedi, G., Hirahara, K., Singleton, K. & O'Shea, J. J. Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Annu. Rev. Immunol. 30, 707–731 (2012).
Hughes, T. et al. Interleukin-1β selectively expands and sustains interleukin-22+ immature human natural killer cells in secondary lymphoid tissue. Immunity 32, 803–814 (2010).
Van de Berg, P. J., van Leeuwen, E. M., ten Berge, I. J. & van Lier, R. Cytotoxic human CD4+ T cells. Curr. Opin. Immunol. 20, 339–343 (2008).
Van Kaer, L. et al. CD8αα+ innate-type lymphocytes in the intestinal epithelium mediate mucosal immunity. Immunity 41, 451–464 (2014).
Karo, J. M., Schatz, D. G. & Sun, J. C. The RAG recombinase dictates functional heterogeneity and cellular fitness in natural killer cells. Cell 159, 94–107 (2014).
Acknowledgements
The authors would like to thank all members of the laboratory for helpful discussions. This work is supported by grants from the Institut Pasteur, Institut National de la Santé et de la Recherche Médicale (INSERM), Ligue National Contre le Cancer and the Agence National pour la Recherche. The authors apologize to those colleagues whose work has not been cited owing to space constraints.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
James P. Di Santo is a founder and stakeholder in AXENIS (Paris, France).
Rights and permissions
About this article
Cite this article
Serafini, N., Vosshenrich, C. & Di Santo, J. Transcriptional regulation of innate lymphoid cell fate. Nat Rev Immunol 15, 415–428 (2015). https://doi.org/10.1038/nri3855
Published:
Issue Date:
DOI: https://doi.org/10.1038/nri3855
This article is cited by
-
Spondyloarthritis with inflammatory bowel disease: the latest on biologic and targeted therapies
Nature Reviews Rheumatology (2023)
-
Whole-genome profiling of DNA methylation and hydroxymethylation identifies distinct regulatory programs among innate lymphocytes
Nature Immunology (2022)
-
Transcriptional control of mature ILC3 function and plasticity: not just RORγt
Cellular & Molecular Immunology (2022)
-
Dietary antigens suppress the proliferation of type 2 innate lymphoid cells by restraining homeostatic IL-25 production
Scientific Reports (2022)
-
Patrolling ILC3 cells monitor intestinal tissue in response to inflammation
Nature Immunology (2022)