Abstract
Surfactant protein D (SFTPD) induces emphysema in knockout mice, but the association of SFTPD with chronic obstructive pulmonary disease (COPD) and emphysema in humans is unclear. Therefore, we aimed to determine the association between genetic variations in SFTPD and susceptibility to COPD and emphysema.
Two populations were studied: population A comprised 270 smokers, including 188 COPD and 82 at-risk subjects, and population B comprised 1131 autopsy cases including 160 cases with emphysema. Six single-nucleotide polymorphisms (SNPs) that tagged the linkage disequilibrium blocks on the entire SFTPD gene were genotyped; the associations of the genotypes with COPD, pulmonary function, percentage of the low-attenuation area (LAA%), and percentage of the airway wall area (WA%) were determined in population A. In population B, the associations of the genotypes with emphysema were assessed.
A C allele at SNP rs721917 that results in the replacement of Met with Thr at position 11 in SFTPD was positively correlated with the LAA% in the upper lung (P=1.1 × 10−5) and overall LAA% (P=1.0 × 10−4), and negatively correlated with the serum concentration of SFTPD (P=7 × 10−11) in the population A. The C/C (rs721917/rs10887199) haplotype was associated with emphysema in both the populations.
Subjects with a C allele at rs721917 have a lower serum SFTPD concentration and are more susceptible to emphysema. This suggests a protective effect of SFTPD against COPD and emphysema.
Similar content being viewed by others
Introduction
Chronic obstructive pulmonary disease (COPD) causes 2.75 million deaths annually and is a serious public health problem.1 COPD is characterized by chronic obstruction of the airflow that is not fully reversible.2 COPD is a heterogeneous disease of which the main phenotypes are emphysema and small airway disease. Each phenotype is considered to develop via a different mechanism.
Tobacco smoking is the most important risk factor for COPD. Nevertheless, only 15–20% of heavy smokers develop symptomatic COPD.3 This suggests that susceptibility to COPD for each individual is, at least in part, genetically determined. Elucidating these genetic factors will clarify the mechanisms of COPD development and therefore the development of emphysema and small airway disease.4
Surfactant protein D (SFTPD) belongs to a group of carbohydrate-binding proteins called collectins and is expressed primarily in type-II alveolar cells.5 SFTPD-knockout mice develop emphysema.6 Moreover, SFTPD concentration in bronchoalveolar fluid is correlated with the airflow limitation in humans.7 These findings indicate that SFTPD is involved in the development of emphysema and that the function of SFTPD may be a determinant of the susceptibility to emphysema. The single-nucleotide polymorphism (SNP) rs721917 affects the amino-acid sequence of SFTPD; two protein isoforms with different functions are derived from this SNP.8, 9 Investigating the association of each isoform with the severity of emphysema will further support the role of SFTPD function in the development of emphysema.
Thus, we designed the current study to examine the association between rs721917 and variables reflecting the severity of emphysema. In addition, we assessed SNPs that are either located inside or flank the SFTPD gene, as the allelic types of such SNPs are often predictive of the expression level of the genes located nearby. Finally, we determined whether SNP allelic types predict the serum concentration of SFTPD and thus susceptibility to emphysema.
Materials and methods
Ethical consideration
The current study was approved by the ethical committees of Nippon Medical School (approval number: 18-11-31), Tokyo Medical and Dental University (approval number: 4), and Tokyo Metropolitan Geriatric Hospital (approval number: 440). Written informed consent was obtained from either the subjects (population A) or from a family member (population B).
Study populations
The characteristics of the two study populations are summarized in Table 1.
Population A: outpatients who were current or ex-smokers had coughing, expectoration, and/or dyspnea, and who had visited the Respiratory Care Clinic, Nippon Medical School for ambulatory treatment between April 2007 and April 2009 were invited to participate in the study.
Population B: subjects who were autopsied at Tokyo Metropolitan Geriatric Hospital between February 1995 and July 2003, who had undergone pathological examination to determine emphysema type and severity, and whose genomic DNA had been preserved for genotyping analysis were included in the study.
Classification of subjects
In population A, subjects who had a diagnosis of COPD according to the Global Initiative for Chronic Obstructive Lung Disease (GOLD) criteria,2 were classified into the COPD group (n=188). The others were classified into the ‘at-risk’ group (n=82). Indicators of emphysema, including the percentage of low-attenuation area (LAA%), LAA% in the upper lung field (LAA%-U), and the percentage of the airway wall area (WA%; an indicator of small airway disease) were measured.
In population B, each subject had undergone pathological examination to determine the emphysema type (centrilobular, panlobular, or focal) and severity (0, none; 1, minimal; 2, mild; 3, moderate; or 4, severe)10, 11 as described in our previous report.12 Briefly, both the excised lungs were inflated with a 10% formalin solution at a constant pressure of 25 cm H2O. Sagittal slices approximately 2 cm thick were obtained. The types and severity of emphysematous changes in the lung slices were macroscopically assessed, and three pathologists (TA, TK, and MS) independently scored them. The scoring was confirmed to be devoid of inter- and intra-observer difference.12 The mean scores of the three pathologists for all slices of both the lungs were used in the current study. Subjects with the centrilobular type with a severity of 3 or 4, and/or the panlobular type with a severity ≥1 were included in the emphysema group. The main phenotypes of emphysema are centrilobular, panlobular, and perilobular. We discounted the perilobular type and only included the centrilobular and panlobular types because centrilobular emphysema is mainly caused by smoking, and panlobular emphysema with centrilobular emphysema is an aggravated form of centrilobular emphysema.13 Subjects without these two types were included in the non-emphysema group.
COPD-related parameters used for the association study
(A) Pulmonary function parameters
Post-bronchodilator-forced expiratory volume in 1 s (FEV1), carbon monoxide-diffusing capacity (diffusing capacity divided by alveolar volume, DLCO/VA), vital capacity (VC), and forced VC (FVC) were measured according to the American Thoracic Society guidelines14 using equipment for lung function testing (CHESTAC; CHEST Co., Tokyo, Japan). We used the reference values of post-bronchodilator FEV1 and VC specified by the Japanese Respiratory Society.15
(B) High-resolution computed tomography (HRCT) parameters
LAA% and LAA%-U reflect the severity of emphysema,16 and WA% reflects the severity of small airway disease.17 We performed helical HRCT scans at 1.25 mm collimation, 0.8 s scan time (rotation time), 120 kV, and 100–600 mA with a Light Speed Pro16 CT scanner (GE Co., Tokyo, Japan). LAA%, LAA%-U, and WA% were calculated as described previously.17, 18, 19 Most Japanese COPD patients have emphysema,20 and about a half of at-risk subjects also have LAA.21 Thus, we investigated the correlation between genetic variations of SFTPD and LAA% in population A to elucidate the association between SFTPD and the extent of emphysema.
Serum SFTPD concentration
Serum SFTPD levels were measured using a sandwich enzyme-linked immunosorbent assay (ELISA) involving monoclonal antibody 7C6 and horseradish peroxidase-conjugated monoclonal antibody 6B2 as described previously.22
SNP selection
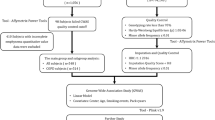
Six tag SNPs that represent most of the genetic variations in SFTPD were selected from the entire SFTPD gene plus a 2-kb 5′ flanking sequence by using Haploview 4.1 software (Whitehead Institute for Biomedical Research, http://www.broad.mit.edu/mpg/haploview) and genotyping data for HapMap JPT (http://www.hapmap.org) (Figure 1). The conditions for the selection were r2>0.8 and a minor allele frequency (MAF)>0.10. Haplotype blocks were also determined by the Haploview software and on the basis of the same genotyping data with a MAF>0.10. The haplotype blocks used were defined according to Gabriel et al.23 We selected two SNPs, rs721917 and rs10887199, for the haplotype analyses as well as genotyping in population B, because these SNPs belong to the same haplotype block (Figure 1) and are associated with COPD (Supplementary Table 1).
The locations of the tag SNPs for the SFTPD gene. A total of six tag SNPs were selected for the entire SFTPD gene and its 5′-flanking sequence (2 kb) using Haploview 4.1 software. The dbSNPs rsID for the tag SNPs are boxed. The strength of the linkage disequilibrium between the two SNPs was measured by r2 (r: correlation coefficient) and is presented as a rhomboid painted by a concordant shade of gray (black: r2=1 and is the strongest; white: r2=0 and is the weakest). Haplotype blocks are delineated by bold lines. The nucleotide number shown at the top is according to the GRCh37 from the Genome Reference Consortium.
Genotyping
Genomic DNA was isolated from the whole blood by using a QIAamp DNA Blood Mini Kit (Qiagen K.K., Tokyo, Japan) (population A) or from the renal cortex by the phenol–chloroform method (population B). SNPs were genotyped by ABI TaqMan SNP Genotyping Assays (Life Technologies Japan, Tokyo, Japan).
Statistical analysis
All values are presented as means and SD or means and SE. Statistical analyses were performed using JMP Genomics software version 3.1 (SAS Institute Inc., Cary, NC, USA). Hardy–Weinberg equilibrium (HWE) was tested for all SNPs in both the populations (Supplementary Table 2). The effects of each SNP genotype on the development of COPD and emphysema were determined using a multivariate logistic regression with dominant, recessive, or additive models on the minor allele. The effect of each SNP genotype on each COPD-related continuous variable or serum SFTPD concentration was calculated using a linear regression model with an additive model. The unobserved haplotype frequencies were estimated by the expectation–maximization (EM) algorithm assuming HWE with 100 iterations. Then, the haplotypes for each subject were speculated as the haplotypes with a maximal probability of >0.95. The effects of each haplotype on each COPD-related parameter or SFTPD concentration were calculated using additive models. In all of these models, the logistic or linear models that maximized either the likelihood or restricted likelihood and thus fit most were considered to be appropriate. Age, gender, smoking status, and pack-years were used as covariates. The odds ratio (OR) of COPD or emphysema development according to the copy number of chromosomal fragments with a specific haplotype was determined using a logistic regression model adjusted for age, sex, smoking status, and pack-years. P-values <0.05 were considered significant. To correct the results for multiple testing, we used a method that accounts for the correlations between tag SNPs and the correlations between clinical phenotypes.24, 25 For six SNPs of SFTPD, an effective number of independent SNPs for correction – 4.82 – was calculated using the Matrix Spectral Decomposition (matSpD) approach26 according to the correlation matrix shown in Supplementary Table 3. By using a similar procedure, we also estimated the effective number of independent phenotypes as 3.98 according to the correlation matrix shown in Supplementary Table 4. Thus, the thresholds of the P-value after correction for the number of SNPs and for the numbers of both SNPs and phenotypes using this method were 0.05/4.82=0.010 and 0.05/(4.82 × 3.98)=0.0026, respectively. T.I. had full access to all of the data in this study and takes responsibility for the integrity of the data and the accuracy of data analysis.
Results
Population A analysis
We found significant associations between COPD and rs721917 (a non-synonymous SNP causing the Met11Thr variation) in a recessive model and between COPD and rs10887199 in a dominant model (Supplementary Table 1). Here, a C allele at rs721917, which results in the Met11Thr variation, and a T allele at rs10887199 were more frequently observed among the patients with COPD than at-risk subjects. SNP rs721917 together with rs2819097 was also significantly associated with LAA%-U (Table 2). Subjects with a C allele at rs721917 were more likely to have a greater LAA% and LAA%-U (Figure 2a). Next, we investigated the association between the rs721917–rs10887199 haplotype and emphysema. There was no significant deviation from HWE in all the SNPs in the subjects of population B without emphysema, namely the control group without emphysema. However, rs721917 deviated from HWE (Supplementary Table 2) in the at-risk subjects, possibly because the at-risk subjects are not disease free (see LAA% in Table 1). Although the EM algorithm assumes HWE, it was recently reported that deviation from HWE does not substantially affect the estimation of haplotypes.27 Haplotype analysis of SNPs rs10887199 and rs721917, which were significantly associated with COPD, showed that the rs10887199 C/rs721917 C haplotype was significantly associated with the presence of COPD and a greater LAA%-U (P=0.029 and 0.014, respectively; OR for COPD=1.58, 95% CI=1.06–2.40) (Table 3). These results indicate that SFTPD is associated with COPD and the pulmonary function parameters of emphysema.
Relationship among rs721917 genotype, LAA%-U, and serum SFTPD concentration. (a) rs721917 genotype and LAA%-U. Each value LAA% indicated is a mean (SD). The number of C allele of rs721917 is positively correlated to LAA%-U. (b) rs721917 genotype and the serum SFTPD concentration (log transformed). The number of C alleles of rs721917 is inversely correlated with the serum SFTPD concentration. (c) LAA%-U and the serum SFTPD concentration. The log-transformed serum SFTPD concentration is inversely correlated with the LAA%-U in subjects who had mild emphysema in population A, namely those with a LAA%-U lower than the median (adjusted P=0.002).
Population B analysis
Population B comprised autopsy cases pathologically diagnosed as either having emphysema (n=160) or not having emphysema (n=971). Although pulmonary function and imaging data for this population were lacking, both DNA samples and pathological data were available, enabling the investigation of the association between SFTPD genotypes and emphysema. We selected two SNPs for genotyping, rs721917 and rs10887199, because they belong to the same haplotype block (Figure 1), are associated with COPD (Supplementary Table 1), and the rs10887199 C/rs721917 C haplotype is associated with COPD and emphysema (Table 3). We did not find any significant association between individual tag SNPs and emphysema (Supplementary Table 5). Nevertheless, the rs721917 C/rs10887199 C haplotype, which was significantly associated with a greater LAA%-U in population A, was also found to be significantly associated with the presence of emphysema (P=0.02; OR=1.45; 95% CI=1.06–1.98; Table 4). These results support the hypothesis that SFTPD is associated with emphysema.
Serum SFTPD concentration
The SNP rs721917 is associated with serum SFTPD concentration in the Caucasians.8, 9 Therefore, we investigated whether a similar association occurs in population A. We found that all tag SNPs, except rs911887 and the rs721917 C/rs10887199 C haplotype (P<1.0 × 10−4; data not shown), were associated with serum SFTPD concentration (Supplementary Table 6, Figure 2b). These results indicate that rs721917 is also associated with the serum SFTPD concentration in the Asians.
A C allele at SNP rs721917 was positively correlated with LAA%-U and negatively correlated with serum SFTPD concentration (Figure 2a and b). This suggests that the association of serum SFTPD concentration and LAA%-U can be observed directly. In severe emphysema, the translocation of SFTPD across the lung–blood barrier increases28, 29 and the serum concentration of SFTPD remains high and unstable due to a high frequency of exacerbations.30 Thus, we divided the patients on the basis of the severity of emphysema and investigated the association between serum SFTPD concentration and emphysema. Because there is no authorized cutoff value for this grouping, we divided the population according to the median value of LAA%-U. As expected, serum SFTPD concentration was inversely correlated with LAA%-U in subjects without severe emphysema in the population A, namely those with an LAA%-U lower than the median (adjusted P-value=0.002, Figure 2c). This again confirms the association between serum SFTPD concentration and emphysema, and demonstrates the advantages of genetic association studies in revealing a correlation between two clinical phenotypes that has not previously been elucidated.
Discussion
The frequency of α1 antitrypsin deficiency is very low among the Japanese.31 Many studies have searched for the genetic factors that determine susceptibility to COPD or emphysema among the Japanese. Nevertheless, only a few factors have been identified. We hypothesized that SFTPD is one of these factors and investigated the associations between the genetic polymorphisms of SFTPD and COPD and emphysema. We found that the Met11Thr variation was associated with emphysema in two independent populations; SFTPD (Thr11) was more frequently found among the emphysema patients. Moreover, patients with a lower serum SFTPD concentration tended to have SFTPD (Thr11) and emphysema.
The association between SFTPD (Thr11) and lower serum SFTPD concentration is consistent with the observation that the Met11Thr polymorphism affects the metabolism of SFTPD protein in humans. The association between serum SFTPD concentration and emphysema suggests that the genetic variations of SFTPD may help develop or progress emphysema via SFTPD concentration in serum and/or epithelial lining fluid. The results of the current study suggest that SFTPD (Thr11) may promote the development of emphysema. Indeed, SFTPD (Thr11) lacks the capacity to form multimers in humans9 and is unable to maintain the homeostasis of surfactant phospholipid in mice.32 These findings suggest that the change from Met11 to Thr11 cripples SFTPD function. However, whether genetic variations of SFTPD also affect emphysema formation by altering its function remains to be elucidated. COPD and emphysema are considered to result from a complicated process, and identifying the main components that determine susceptibility to them is not easy. Nevertheless, the association between SFTPD (Thr11) and both COPD and emphysema together with the fact that SFTPD (Thr11) exhibits altered function and metabolism strongly supports the hypothesis that SFTPD is one of the determinants of COPD and emphysema.
Recently, it was reported that SNPs in SFTPD are associated with COPD in multiple Caucasian populations33 and that SFTPD (Thr11) promotes susceptibility to COPD. These results are consistent with those of our study among the Japanese. Furthermore, these consistent results strengthen the conclusion that SFTPD is involved in the development of COPD and emphysema. An SNP in the promoter region (rs1885551) and an SNP that has a strong linkage disequilibrium with the promoter region (rs10887199; Figure 1) have been shown to be more strongly associated with the serum SFTPD concentration than rs721917 among Caucasians and the Japanese, respectively. Therefore, it is speculated that the promoter region of SFTPD may also affect serum concentration. The finding that serum SFTPD concentration is inversely correlated with emphysema in the upper lungs in subjects without severe emphysema is unique to our study.
Our study has some limitations. Our study population is not very large; the two populations that we investigated (populations A and B) had related pulmonary lesions but were collected in different settings. This inevitably imposes limitations in the interpretation of the study’s results. However, the association between genetic variations in SFTPD is reported in the Caucasian populations33 and was also observed in this study with our Japanese populations. Thus, this association was established among different races and strongly supports the hypothesis that SFTPD is involved in the development of emphysema. In addition, it is an advantage for genetic association studies that our Japanese populations exhibit much less stratification than the Caucasian populations.34 We conclude that genetic variations of SFTPD are strongly related to the development of emphysema.
In conclusion, a C allele at SNP rs721917, which results in the substitution of Met with Thr at position 11 in SFTPD, is related to susceptibility to emphysema. Serum SFTPD concentration predicts SFTPD genotypes and indicates susceptibility to emphysema. The role that SFTPD has in the development of COPD should be investigated further from both pathogenic and therapeutic perspective.
References
Murray CJ, Lopez AD : Evidence-based health policy—lessons from the global burden of disease study. Science 1996; 274: 740–743.
Global strategy for diagnosis, management, and prevention of COPD, http://www.goldcopd.com/ (accessed 13 April 2009).
Snider GL : Chronic obstructive pulmonary disease: risk factors, pathophysiology and pathogenesis. Annu Rev Med 1989; 40: 411–429.
Patel BD, Coxson HO, Pillai SG et al: International COPD genetics network. Airway wall thickening and emphysema show independent familial aggregation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2008; 178: 500–505.
Hartl D, Griese M : Surfactant protein D in human lung diseases. Eur J Clin Invest 2006; 36: 423–435.
Wert SE, Yoshida M, LeVine AM et al: Increased metalloproteinase activity, oxidant production, and emphysema in surfactant protein D gene-inactivated mice. Proc Natl Acad Sci USA 2000; 97: 5972–5977.
Sin DD, Man SF, McWilliams A et al: Surfactant protein D and bronchial dysplasia in smokers at high risk of lung cancer. Chest 2008; 134: 582–588.
Sorensen GL, Hjelmborg JB, Kyvik KO et al: Genetic and environmental influences of surfactant protein D serum levels. Am J Physiol Lung Cell Mol Physiol 2006; 290: L1010–L1017.
Leth-Larsen R, Garred P, Jensenius H et al: A common polymorphism in the SFTPD gene influences assembly, function, and concentration of surfactant protein D. J Immunol 2005; 174: 1532–1538.
Report of a National Heart, Lung, and Blood Institute: Division of Lung Diseases workshop: the definition of emphysema. Am Rev Respir Dis 1985; 132: 182–185.
Yamanaka A : Morphopathology of chronic pulmonary emphysema. Acta Pathol Jpn 1965; 15: 33–39.
Fujimoto K, Ikeda S, Arai T et al: Polymorphism of SERPINE2 gene is associated with pulmonary emphysema in consecutive autopsy cases. BMC Med Genet 2010; 11: 159–165.
Chapter 4. Emphysema: Classification, Morphology, and Associations,; In: Wright JL and Thurlbeck WM (eds): Thurlbeck's Chronic Airflow Obstruction. Second edition. Hamilton, Ontario, B.C. Decker, Inc., 1999, pp 85–144.
American Thoracic Society: Standardization of spirometry, 1994 update. Am J Respir Crit Care Med 1995; 152: 1107–1136.
Japanese Respiratory Society: The predicted values of spirometry and arterial blood gas analysis in Japanese. J Jap Resp Soc 2001; 39: Appendix (in Japanese).
Anderson Jr AE, Foraker AG : Centrilobular emphysema and panlobular emphysema: two different diseases. Thorax 1973; 28: 547–550.
Nakano Y, Muro S, Sakai H et al: Computed tomographic measurements of airway dimensions and emphysema in smokers. Correlation with lung function. Am J Respir Crit Care Med 2000; 162: 1102–1108.
Okazawa M, Müller N, McNamara AE et al: Human airway narrowing measured using high resolution computed tomography. Am J Respir Crit Care Med 1996; 154: 1557–1562.
Orlandi I, Moroni C, Camiciottoli G et al: Chronic obstructive pulmonary disease: thin-section CT measurement of airway wall thickness and lung attenuation. Radiology 2005; 234: 604–610.
Tatsumi K, Kasahara Y, Kurosu K et al: Respiratory Failure Research Group in Japan: Clinical phenotypes of COPD: results of a Japanese epidemiological survey. Respirology 2004; 9: 331–336.
Betsuyaku T, Yoshioka A, Nishimura M et al: Pulmonary function is diminished in older asymptomatic smokers and ex-smokers with low attenuation areas on high-resolution computed tomography. Respiration 1996; 63: 333–338.
Inoue T, Matsuura E, Nagata A et al: Enzyme-linked immunosorbent assay for human pulmonary surfactant protein D. J Immunol Methods 1994; 173: 157–164.
Gabriel SB, Schaffner SF, Nguyen H et al: The structure of haplotype blocks in the human genome. Science 2002; 296: 2225–2229.
Nyholt DR : A simple correction for multiple testing for single nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet 2004; 74: 765–769.
Daley D, Lemire M, Akhabir L et al: Analyses of associations with asthma in four asthma population samples from Canada and Australia. Hum Genet 2009; 125: 445–459.
Matrix Spectral Decomposition (matSpD), estimate the equivalent number of independent variables in a correlation (r) matrix http://gump.qimr.edu.au/general/daleN/matSpD/ accessed in May 2011.
Fallin D, Schork NJ : Accuracy of haplotype frequency estimation for biallelic loci, via the expectation-maximization algorithm for unphased diploid genotype data. Am J Hum Genet 2000; 67: 947–959.
Antonelli Incalzi R, Maini CL et al: 99mTc-DTPA clearance in bullous emphysema. Nuklearmedizin 1985; 24: 180–184.
Tkacova R, McWilliams A, Lam S et al: Integrating lung and plasma expression of pneumo-proteins in developing biomarkers in COPD: a case study of surfactant protein D. Med Sci Monit 2010; 16: CR540–CR544.
Lomas DA, Silverman EK, Edwards LD et al: Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints study investigators. Serum surfactant protein D is steroid sensitive and associated with exacerbations of COPD. Eur Respir J 2009; 34: 95–102.
Saito A, Takizawa H, Sato M et al: Alpha-1 antitrypsin deficiency with severe pulmonary emphysema. Intern Med 2004; 43: 223–226.
Zhang L, Hartshorn KL, Crouch EC, Ikegami M, Whitsett JA : Complementation of pulmonary abnormalities in SP-D(-/-) mice with an SP-D/conglutinin fusion protein. J Biol Chem 2002; 277: 22453–22459.
Foreman MG, Kong X, Demeo DL et al: Polymorphisms in surfactant protein-D are associated with chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol 2011; 44: 316–322.
Yamaguchi-Kabata Y, Nakazono K, Takahashi A et al: Japanese population structure, based on SNP genotypes from 7003 individuals compared to other ethnic groups: effects on population-based association studies. Am J Hum Genet 2008; 83: 445–456.
Acknowledgements
We sincerely thank Ms Fujishiro for helping with our research. We also acknowledge Drs Kurosaki H, Motohashi N, Morii K, Hattori K, Yamada K, and Motegi T, our nurses and other colleagues, and the patients involved in the study for the collection of the samples. Editage checked this manuscript to ensure an acceptable standard of English. This study was partly supported by a grant from the Grant-in-Aid for Young Scientists (B) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflicts of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Ishii, T., Hagiwara, K., Kamio, K. et al. Involvement of surfactant protein D in emphysema revealed by genetic association study. Eur J Hum Genet 20, 230–235 (2012). https://doi.org/10.1038/ejhg.2011.183
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2011.183
Keywords
This article is cited by
-
Surfactant protein D, Club cell protein 16, Pulmonary and activation-regulated chemokine, C-reactive protein, and Fibrinogen biomarker variation in chronic obstructive lung disease
Respiratory Research (2014)
-
Differences in serum SP-D levels between German and Japanese subjects are associated with SFTPDgene polymorphisms
BMC Medical Genetics (2014)
-
Genetic Polymorphisms of Surfactant Protein D rs2243639, Interleukin (IL)-1β rs16944 and IL-1RN rs2234663 in Chronic Obstructive Pulmonary Disease, Healthy Smokers, and Non-Smokers
Molecular Diagnosis & Therapy (2014)