Abstract
Interferon regulatory factor-4 binding protein (IBP) is a novel upstream activator of Rho GTPases. Our previous studies have shown that ectopic expression of IBP was correlated with malignant behaviors of human breast cancer cells, and invasive human breast cancer had high expression of IBP that promoted the proliferation of these cells. However, it remains unknown whether autophagy inhibition contributes to IBP-mediated tumorigenesis. In this study, we for the first time, reported that upregulation of IBP expression significantly suppressed the autophagy of breast cancer cells, and downregulation of IBP expression markedly induced autophagy of these cells. Further investigation revealed that IBP effectively counteracted autophagy by directly activating mammalian target of rapamycin complex 2 (mTORC2) and upregulating phosphorylation of Akt on ser473 and FOXO3a on Thr32. Moreover, IBP-mediated suppression of autophagy was dependent on mTORC2/Akt/FOXO3a signaling pathway. Finally, our results demonstrated that IBP-mediated breast cancer cell growth in vitro and in vivo was strongly correlated with suppression of mTORC2-dependent autophagy. These findings suggest that the anti-autophagic property of IBP has an important role in IBP-mediated tumorigenesis, and IBP may serve as an attractive target for treatment of breast cancer.
Similar content being viewed by others
Main
Breast cancer remains a leading cause of morbidity and mortality in women.1 Although more and more women survive from breast cancer due to early detection, ∼30% patients who are diagnosed with breast cancer at an early stage eventually develop recurrence or metastasis.2 Therefore, to understand the pathophysiology of breast cancer and to investigate new therapeutic targets are critical for the development of novel therapeutic strategies to improve the clinical outcome.
Interferon regulatory factor binding protein (IBP) was first identified in 20033 and has been found to have multiple roles in the biological processes of the immune system. Our previous work found IBP was highly expressed in invasive human breast cancer.4 Moreover, overexpression of wild-type IBP in an IBP-negative breast cancer cell line markedly increased its proliferation and invasiveness in vitro. In contrast, RNA interference-mediated knockdown of IBP expression in an IBP-positive breast cancer cell line significantly reduced the growth and invasiveness of these cells.4 These results indicate that IBP has an important role in the proliferation and metastasis of breast cancer cells, and may serve as a novel tumor marker and therapeutic target for breast cancer. However, the exact mechanisms of IBP-mediated tumorigenesis in breast cancer are still unclear.
Autophagy is a major intracellular pathway for degradation and recycling of proteins, ribosomes and entire organelles.5 Increasing evidence suggests that autophagy may be a cell death mechanism under certain cellular scenarios, and this autophagy-dependent cell death has been defined as ‘autophagic cell death’ or ‘type II programmed cell death.’6 It has also been suggested that alterations in the pathways regulating autophagic cell death may result in cancer development.7, 8, 9 Knocking out or inactivation of autophagy-related genes results in increased tumorigenesis in mammal models, and overexpression of these genes suppresses the formation of various human cancers such as breast, ovarian and prostate cancer.7, 10, 11 Recent studies demonstrate that autophagy defects in cancers cause accumulation of unwanted proteins (such as p62 and ER chaperons), oxidative stress and genomic damage, all of which concomitantly fuel cancer growth.12, 13 Importantly, Oh et al.14 reported that downregulation of autophagy by Bcl-2 promoted MCF-7 breast cancer cell growth independent of inhibiting apoptosis. The above findings indicate that autophagy is closely related to the formation of breast cancer. However, it is unclear whether IBP exerts effects in breast cancer by regulating autophagy. Mavrakis et al.15 found that stimulation of the phosphatidylinositol 3-kinase(PI3K) signal pathway resulted in translocation of an overexpressed IBP–GFP fusion protein to cell membrane, suggesting that IBP may exert its effects by affecting the PI3K signal pathway. Importantly, it has been known that the PI3K/Akt/ mammalian target of rapamycin (mTOR) ((PI3K/Akt/mTOR)) pathway has a decisive role in the negative regulation of autophagy. The inhibitors of class I PI3K, Akt and mTOR can inhibit this cascade and then induce autophagy.16, 17 Moreover, the PI3K/Akt/mTOR signaling pathway has critical roles in neoplastic cell growth.18, 19 Thus, we postulate that IBP may promote the growth and metastasis of breast cancer cells by regulating autophagy in a PI3K/Akt/mTOR signaling pathway-dependent manner.
In the PI3K/Akt/mTOR signaling cascade, mTOR has been shown to participate in two signaling complexes with distinct cellular functions: mTORC1 and mTORC2.20 mTORC1 is transformed from mTOR, raptor and mLST8; whereas, mTORC2 includes mTOR, rictor, mLST8 and SIN1.21 mTORC2 as a regulatory Ser473 kinase of Akt has an important regulatory role in growth factor signaling by regulating PI3K and its crucial downstream effector Akt.22 Recent studies found that FoxO3 transcription factor is necessary and sufficient for the induction of autophagy. FOXO3a belongs to the Forkhead family of transcription factors: FKHRL or FOXO. The phosphorylation in Thr32 of FOXO3a results in translocation of FOXO from the nucleus, which inhibits the transcription of autophagy-related genes.23, 24 Importantly, Akt activation of Ser473, a downstream target of mTORC2, promotes the FoxO3 phosphorylation, and autophagy is suppressed accordingly. This effect is not prevented by rapamycin (a mTORC1 inhibitor). Thus, the mTORC2/Akt/FOXO3a signaling pathway has a crucial role in the suppression of autophagy in many cellular settings.
In this study, the IBP-suppressed autophagy was investigated in human breast cancer cells in vitro and in vivo. The mechanisms underlying autophagy inhibition and cell growth and metastasis induced by IBP were explored in human breast cancer cells. Our in vitro and in vivo findings revealed that OSI-027 (a mTORC2/mTORC1 inhibitor) not only reversed the IBP-mediated autophagy inhibition, but also obviously blocked the IBP-mediated activation of the mTORC2/Akt/FOXO3a signaling pathway, and then inhibited the IBP-induced cell growth and metastasis of breast cancer cells. Thus, our study for the first time indicates that the anti-autophagic property of IBP has a key role in the IBP-mediated tumorigenesis, and may, to some extent, serve as an attractive target for breast cancer therapy.
Results
IBP is an inhibitor of autophagy in breast cancer
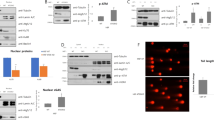
MDA-MB-231 cells positive for IBP expression and MDA-MB-468 cells negative for IBP expression were used in the in vitro experiments (Figure 1a). Western blot assay showed that MDA-MB-231 cells treated with two different IBP siRNAs recombinant plasmid #1 and #2 had 57–86% reduction in the IBP expression (Figure 1b). Consistently, overexpression of IBP in MDA-MB-468 cells was found after transfection with pEGFP–IBP expression plasmid (Figure 1c). In autophagy, LC3-I is converted to lipidated LC3-II, which is the classical hallmarks of autophagy.25, 26 p62 protein is a well-known autophagic substrate.27, 28 To address the role of IBP in autophagy of breast cancer cells, autophagy was detected with three classical methods. First, we found that IBP suppressed the conversion of LC3-II, leading to an increased expression of p62 after starvation-induced stimulation. Figure 1d shows significant increase in the ratio of LC3-II to actin and notable decline in p62 in MDA-MB-231-IBP RNAi cells. Similar results were observed in MDA-MB-468 cells (Figure 1e). Furthermore, autophagic flux was monitored in the presence of lysosomal protease inhibitors, bafilomycin, in basal and starvation conditions.26, 29 As shown in Figures 1f and g, bafilomycin enhanced the accumulation of LC3-II, indicating that autophagic flux was intact and supraphysiological autophagic response was indeed induced by IBP knockdown. In addition, we found that suppression of IBP expression markedly increased the accumulation of RFP-LC3-positive vesicles in MDA-MB-231-IBP RNAi cells (Figure 1h). Finally, an extensive accumulation of autophagosomes was observed in MDA-MB-231-IBP RNAi cells. Conversely, reduced accumulation of autophagosomes was observed in MDA-MB-468-IBP cells (Figures 1i and j). The above results convincingly demonstrate that IBP is an inhibitor of autophagy in breast cancer cells.
IBP-inhibited autophagy in breast cancer cells. (a) Western blot analysis for IBP in breast cancer lines including MDA-MB-231 and MDA-MB-468 cells. (b) Detection of stable inhibitory efficiency of siRNAs against IBP in MDA-MB-231cells. (c) Immunoblotting detected construction of IBP overexpression model in MDA-MB-468 cells. (d) Control siRNA and siIBP MDA-MB-231 breast cancer cells were cultured in basal conditions or starved for 0.5, 2, 3 h with HBSS. The inhibition of IBP expression enhanced LC3-II level and promoted the degradation of p62 in MDA-MB-231 cells. (e) Control MDA-MB-468 and MDA-MB-468-IBP breast cancer cells were cultured in basal conditions or starved for 1, 2, 3 h with HBSS. Overexpression of IBP inhibited LC3-II level and reduced the degradation of p62 in MDA-MB-468 cells. (f and g) Autophagic flux analysis showed bafilomycin enhanced the accumulation of LC3-II. The above breast cancer cells were incubated with or without bafilomycin (10 nM) in basal conditions and autophagy induction by HBSS starvation stimulation for 6 h. (h) IBP-inhibited autophagy in breast cancer cells as analyzed by RFP-LC3 puncta formation using a laser scanning confocal microscopy. The percentage of cells with RFP-LC3 punctate was quantified in 50 red fluorescent cells in one field. (i and j) Representative electron microscopic images show autophagic vacuoles in breast cells. The percentage of cells with autophagic vacuoles was quantified in 20 cells each group. **P<0.01 versus control cell group
IBP activates the mTORC1 and mTORC2 signaling pathway in breast cancer cell lines
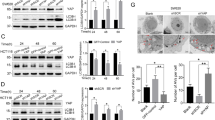
To examine the mechanism of IBP-mediated autophagy inhibition, the effects of IBP on the PI3K/Akt/mTOR pathway were observed.30 Results showed that IBP mainly upregulated the mRNA expressions of PI3K, Akt, rictor and mTOR in breast cancer cells (Figures 2a and b). Recent studies demonstrate that mTORC1 contains primarily Ser2448 phosphorylation, whereas mTORC2 presents with Ser2481 phosphorylation, and Ser2481 phosphorylation of mTOR is a marker for the presence of mTORC2 complexes.31 As shown in Figure 2c, IBP knockdown in MDA-MB-231 cells markedly inhibited p-mTOR Ser2481, and IBP overexpression in MDA-MB-468 cells promoted p-mTOR Ser2481. In addition, direct mTORC2 substrate p-Akt Ser473 was markedly inhibited in MDA-MB-231-IBP siRNA cells, but promoted in MDA-MB-468-IBP cells. Similar results were observed on p-FOXO3a Thr32.
IBP activated the mTORC1 and mTORC2 signaling pathway in breast cancer cells. (a and b) qRT-PCR analysis of the mTOR signaling pathway-associated gene expression levels in MDA-MB-231 and MDA-MB-468 cells with different levels of IBP. (c) Those cells were analyzed for mTORC2 activity by immunoblotting for levels of p-mTOR Ser2481, p-Akt Ser473 and p-FOXO3a Thr32. (d) Cells were analyzed for mTORC1 activity by immunoblotting for levels of p-mTOR Ser2448, p-P70S6K Thr389 and p-4EBP1 Thr37/46
In addition, as shown in Figure 2d, the knockdown of IBP in MDA-MB-231 cells markedly inhibited mTORC1 pathway: p-mTOR Ser2448, p-P70S6K Thr389 and p-4EBP1 Thr37/46. Conversely, IBP overexpression in MDA-MB-468 cells promoted p-mTOR Ser2448, p-p70S6K Thr389 and p-4EBP1 Thr37/46.
IBP-mediated autophagy inhibition is dependent on the mTORC2 signaling pathway in breast cancer cells
It has been known that PI3K is an upstream component of mTORC1 and mTORC2 pathways and its downstream targets include Akt and mTOR, resulting in the inactivation of autophagy.32, 33 To further confirm which signaling pathway the IBP-mediated inhibition of autophagy depends on, some specific inhibitors such as PI3K inhibitor-LY294002, mTORC1 inhibitor-rapamycin and mTORC2/mTORC1 inhibitor-OSI-027 were used. As shown in Figures 3a and b, only OSI-027 could significantly reverse the IBP-mediated inhibition of LC3-II/actin ratio and p62 degradation. In addition, as shown in Figures 3c and d, OSI-027 not only abolished the IBP-mediated reduction in LC3-II/actin ratio, but reversed the IBP-mediated activation of p-mTOR Ser2481 and its downstream targets in breast cancer cells. Moreover, rapamycin together with OSI-027 showed similar effects as OSI-027 alone did. Therefore, the IBP-mediated inhibition of autophagy is mainly dependent on the mTORC2 signaling pathway.
IBP-mediated inhibition of autophagy is dependent on the mTORC2 signaling pathway in breast cancer cells. (a and b) MDA-MB-231 and MDA-MB-468 cells with different levels of IBP were treated without or with LY294002 (25 μm), rapamycin (10nm), OSI-027 (10 μm). Immunoblotting detected the levels of autophagy. (c and d) Cells were incubated with either rapamycin (10 nm) or OSI-027 (10 μm) or combined the two for 2 h. Immunoblotting detected the level of p-mTOR Ser2481, p-Akt Ser473, p-FOXO3a Thr32 and LC3-II
mTORC2 pathway-dependent autophagy is implicated in IBP-induced growth and survival of breast cancer cells
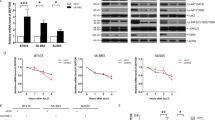
MTT assay showed that suppression of IBP expression markedly reduced the growth and survival of MDA-MB-231- IBP siRNA cells, whereas IBP overexpression promoted the growth and survival of MDA-MB-468-IBP cells (Figures 4a and b). Moreover, OSI-027 obviously blocked the IBP-induced growth and survival of both MDA-MB-231-control siRNA cells and MDA-MB-468-IBP cells. Although rapamycin inhibited the IBP-induced growth and survival of breast cancer cells, its effect was weaker than OSI-027 did. Moreover, OSI-027 together with rapamycin failed to enhance the IBP-induced growth and survival of breast cancer cells. In addition, only OSI-027 could block the IBP-induced colony formation (Figures 4c and d).
mTORC2 pathway-dependent inhibition of autophagy is implicated in IBP-induced breast cancer cells growth and survival. (a and b) MDA-MB-231 and MDA-MB-468 cells with different levels of IBP were treated with either rapamycin (10 nm) or OSI-027 (10 μm) or combined the two for 48 h. MTT-based cell viability assay was performed in breast cancer cells (**P<0.01 versus control-siRNA; #P<0.05 and ##P<0.01 versus DMSO-treated control-siRNA cells in a). (**P<0.01 versus EGFP-IBP; #P<0.05 and ##P<0.01 versus DMSO-treated EGFP-IBP cells in b). (c and d) Cells were treated with or without rapamycin (10 nM), OSI-027 (10 μM) or both for 2 weeks. Clonogenic survival assay was performed (**P<0.01 versus control-siRNA; #P<0.05 and ##P<0.01 versus DMSO-treated control-siRNA cells). Data are the mean±S.D. of three different experiments
mTORC2 pathway-dependent autophagy is implicated in IBP-induced migration and invasiveness of breast cancer cells
Wound-healing assay (Figures 5a and b) and Transwell invasion assay (Figures 5c–e) showed that IBP knockdown in MDA-MB-231 cells inhibited their migration and invasiveness, but IBP overexpression in MDA-MB-468 cells promoted their migration and invasiveness. Moreover, only OSI-027 significantly inhibited the IBP-induced migration and invasiveness. Importantly, OSI-027 together with rapamycin failed to enhance the effects of OSI-027.
mTORC2 pathway-dependent inhibition of autophagy is implicated in IBP-induced breast cancer cells migration and invasiveness. (a and c) MDA-MB-231 and MDA-MB-468 cells with different levels of IBP were treated with either rapamycin (10 nm) or OSI-027 (10 μm) or combined the two for 48 h. Wound coverage and invasion of cells were then monitored. (b) Quantitative presentation of wound coverage was performed (**P<0.01 versus control-siRNA; #P<0.05 and ##P<0.01 versus DMSO-treated control-siRNA cells). (d and e) Data show the amount of invasion of cells (**P<0.01 versus control-siRNA; #P<0.05 and ##P<0.01 versus DMSO-treated control-siRNA cells) (**P<0.01 versus EGFP-IBP; #P<0.05 and ##P<0.01 versus DMSO-treated EGFP-IBP cells). Data are the mean±S.D. of three different experiments
Inhibition of autophagy via mTORC2-dependent signal pathway is required for IBP-mediated growth and metastasis of human breast cancer cells in vivo
As shown in Figure 6a, tumors from control siRNA mice showed decreased LC3-II conversion together with activated mTORC2 pathway. A vast majority of tumor cells from the siIBP mice showed a large amount of membranous vacuoles resembling autophagosomes (Figures 6b and c). As shown in Figure 6d, the tumor growth in the siIBP group was significantly inhibited. Only OSI-027 significantly blocked the IBP-induced growth of xenografts. OSI-027 together with rapamycin failed to enhance the effects of OSI-027 on IBP-induced xenograft growth. Surprisingly, as shown in Figures 6e–h, more tumor foci from lung and liver were found in the IBP-implanted mice than the siIBP-implanted mice. Furthermore, we found that OSI-027 alone significantly blocked the IBP-induced lung and liver metastasis, and OSI-027 together with rapamycin did not enhance the above effects of OSI-027 alone used.
IBP facilitated MDA-MB-231 xenograft tumor growth and metastasis in vivo in association with mTORC2 pathway-dependent inhibition of autophagy. MDA-MB-231 cells with different levels of IBP (9 × 106) were s.c. injected into the flanks on right side of each mouse. (a) IBP-inhibited autophagy in MDA-MB-231 xenograft tumors in vivo through the mTORC2 pathway. (b) Autophagic vacuoles were shown by transmission electron microscopy in a xenograft model. (c) The percentage of cells with autophagic vacuoles was quantified in 20 cells per group. (**P<0.01 versus control group) (d) IBP potently promoted the MDA-MB-231 xenograft tumor growth by activating the mTORC2 pathway. (e and f) The number of lung and liver metastases was counted and plotted (n=7) between these groups. (g) Images of representative liver metastasis and lung metastasis were shown. (h) The main organs of mice between these groups were visualized by H&E staining
Inhibited autophagy by IBP overexpression and activation of mTORC2 pathway are observed in breast cancer patients
As shown in Figure 7a and Table 1, IBP expression was higher in breast cancer tissues than that in the adjacent normal tissues (P<0.001). This is in line with our previous results.4 Besides, the p-mTOR S2481-positive rate significantly increased in breast cancer tissues. Conversely, the LC3-II positive rate significantly reduced in breast cancer tissues (0%) when compared with adjacent normal tissues (60%) (Figure 7a). Correlation analysis showed there was a negative relationship between IBP and LC3-II protein levels (P<0.001), but a significant positive correlation (P<0.001) between IBP expression and p-mTOR on Ser2481 (Table 1). As shown in Figure 7b and c, IBP expression and the levels of p-mTOR on Ser2481, p-Akt on S473 and p-FOXO3a on T32 was markedly higher in breast cancer tissues than in the adjacent normal tissues. Conversely, LC3-II expression and the autophagosomes formation were lower in breast cancer tissues than in adjacent normal tissues.
IBP-suppressed autophagy through activation of mTORC2 pathway in human breast cancer tissues. (a) Expression of IBP, p- mTOR (Ser2481) and LC3II was assessed in breast cancer tissue microarray by immunohistochemistry. (b) Paired breast tumor and matched normal adjacent tissue lysates from the same patient donors was used to detect the level of mTORC2 pathway and autophagy by immunohistochemistry. (c) Representative electron microscopic images showed autophagic vacuoles in paired breast tumor and matched normal adjacent tissue lysates from the same patient donors
Discussion
In this study, we found that IBP-positive rate significantly increased in breast cancer tissues, which was in line with our previous results.4 However, the mechanisms of IBP-mediated tumorigenesis remain largely unknown. In this study, we report for the first time that IBP inhibits autophagy of breast cancer cells. First, our in vitro experiments showed that IBP markedly suppressed the conversion of LC3-I to LC3-II, accumulation of RFP-LC3-positive vesicles and formation of autophagosomes, leading to an increased expression of p62 protein at different time points after starvation-induced stimulation with HBSS.34 In addition, in IBP-induced growth and metastasis of xenografts in vivo, results showed tumors of mice after inoculation of MDA-MB-231- IBP siRNA cells presented with increased LC3-II conversion and autophagosomes formation. Moreover, all breast cancer tissues (with high IBP expression) had no LC3 protein expression, but nearly 60% of adjacent normal tissues (without IBP expression) were positive for LC3 protein expression. Our findings thus indicate that IBP is an inhibitor of autophagy in breast cancer. It has been known that autophagy is a key mechanism in various physiopathological processes, including tumorigenesis, development, cell death and survival.35, 36 Importantly, autophagy also has an important role in the growth, migration and invasiveness of breast cancer cells. Akar et al.37 provided the evidence that targeted silencing of Bcl-2 induced autophagic cell death in MCF-7 breast cancer cells. Indelicato et al.38 found that TFP (an activator of autophagy) inhibited the invasion, migration and survival of breast cancer cells. Chiavarina et al.39 found that HIF1-α functioned as a tumor suppressor by activating autophagy of MDA-MB-231 breast cancer cells to repress tumor growth via the ‘self-digestion.’ These findings suggest that autophagy serves as a pro-death mechanism in breast cancer cells. Thus, we hypothesize that IBP-mediated autophagy inhibition may represent an important mechanism of tumorigenesis in breast cancer. However, an important question arises that why IBP possesses the ability to inhibit autophagy and how the IBP-mediated deregulation of autophagy contributes to tumor development.
It has been known that the PI3K/Akt/mTOR signaling pathway has a decisive role in the negative regulation of autophagy.30 In this study, our findings indicated that IBP mainly upregulated the mRNA expression of PI3K, Akt and mTOR in breast cancer cells, suggesting that IBP may inhibit autophagy via activating the PI3K/Akt/mTOR signaling pathway. Recent studies showed that the autophagy stimulation in many cellular settings relies on the inhibition of mTOR complex, which has a central role in controlling protein synthesis, cell growth and cell proliferation through regulating several downstream targets.40 Based on their sensitivity to rapamycin, mTOR nucleates two distinct multiprotein complexes, mTORC1 and mTORC2.41, 42 mTORC1 contains primarily p-mTOR on Ser2448, whereas mTORC2 contains p-mTOR on Ser2481.31 More recently, studies have been conducted to investigate the role of mTORC2 in autophagy. Carayol et al.43 reported that mTORC2/mTORC1 inhibitor OSI-027 could induce autophagic cell death in BCR-ABL-expressing leukemic cells. Moreover, mTORC2 kinase as a major hydrophobic kinase can phosphorylate Akt on Ser473,44, 45 which means that Akt phosphorylation on S473 is due to the mTORC2 kinase activation. Importantly, recent studies showed that FoxO3a transcription factor is necessary and sufficient for the induction of autophagy. This effect of FoxO3a cannot be prevented by rapamycin (an mTORC1 inhibitor).23, 24 It is indicated that the mTORC2/Akt/FOXO3a signaling pathway has a crucial role in autophagy suppression in many cellular settings. In addition, some studies show that mTORC1 complex is sensitive to rapamycin and also negatively regulated autophagy by phosphorylating p70S6K and 4E-BP1.20, 46, 47
In order to examine the exact mechanism of IBP-mediated inhibition of autophagy, we observed the effects of IBP on the mTORC2/Akt/FOXO3a and the mTORC1 signaling pathway. Our results showed that IBP obviously promoted the phosphorylations of mTOR on Ser2481, Akt on Ser473 and FOXO3a on Thr32 in breast cancer cells after starvation-induced stimulation, indicating that IBP can activate mTORC2/Akt/FOXO3a signaling pathway. Here, the detail mechanism about how does IBP activate mTORC2/Akt/FOXO3a signaling pathway in breast cancer cells is still unclear. At the same time, we also found that IBP could activate mTORC1 signaling pathway. Thus, whether IBP inhibits autophagy via activating the mTORC2/Akt/FOXO3a signaling pathway alone or concurrently activating the mTORC1 signaling pathway is still unclear. Here, in order to elucidate the question, three different selective inhibitors of the PI3K/Akt/mTOR signaling pathway (LY294002 (a PI3K inhibitor), rapamycin (an allosteric inhibitor of mTORC1) and OSI-027 (a potent inhibitor of mTORC2 and mTORC1)) were used.48Interestingly, we found only OSI-027 reversed the IBP-mediated inhibition of autophagy, compared with rapamycin and LY294002. Moreover, we found that OSI-027 also blocked the IBP-mediated activation of the mTORC2/Akt/FOXO3a signaling pathway in breast cancer cells, but OSI-027 together with rapamycin did not enhance the effect of OSI-027 alone used. In addition, in IBP-induced xenografts model in vivo, we also found IBP decreased autophagy together with activated mTORC2/Akt/FOXO3a pathway in breast cancer tissues. Thus, the above results indicate that IBP-mediated inhibition of autophagy is dependent on the mTORC2/Akt/FOXO3a signaling pathway.
Interestingly, we want to know whether mTORC2/Akt/FOXO3a pathway-dependent autophagy inhibition is implicated in IBP-induced growth and metastasis of breast cancer cells. To elucidate the question, in vitro experiments and in vivo xenografts model were used. First, in MTT and clonogenic survival assay, we found that IBP markedly promoted the growth and survival of breast cancer cells. Only OSI-027 significantly blocked the IBP-induced growth and survival of the breast cancer cells and OSI-027 together with rapamycin did not show synergistic effect. This demonstrates that IBP-medicated autophagy inhibition via mTORC2/Akt/FOXO3a pathway is essential for IBP-induced promotion of growth and survival of breast cancer cells. Second, in wound healing and Transwell invasion assay, we also found IBP obviously promoted the migration and invasiveness of breast cancer cells. Compared with rapamycin group, OSI-027 significantly blocked the IBP-induced increase of migration and invasiveness of breast cancer cells, but OSI-027 together with rapamycin failed to enhance the effects of OSI-027 alone used. These findings thus indicate that the IBP-mediated inhibition of autophagy via the mTORC2-dependent signal pathway is also essential for IBP-induced increase of migration and invasiveness of breast cancer cells. Third, in in vivo xenografts, we found that IBP markedly promoted the growth of MDA-MB-231-control siRNA xenografts and induced the lung and liver metastasis. Only OSI-027 significantly blocked the IBP-induced growth of xenografts and the IBP-induced lung and liver metastasis. OSI-027 together with rapamycin failed to enhance these effects of OSI-027. Therefore, our in vivo and in vitro studies demonstrate that the activation of the mTORC2 pathway is necessary for IBP-dependent autophagy suppression and the subsequent promotion of growth and metastasis of breast cancer cells. These effects of IBP are not related to the mTORC1 pathway.
To further understand the physiological significance of IBP-mediated inhibition of autophagy in a mTORC2 pathway-dependent manner in breast cancer, breast cancer and adjacent normal tissues from breast cancer patients were used to observe the effects of IBP expression on autophagy and the mTORC2/Akt/FOXO3a signaling pathway. Surprisingly, the same results are observed. We found that both the expression of IBP and mTORC2/Akt/FOXO3a pathway-related molecules were markedly higher in breast cancer tissues than that in adjacent normal tissues. Conversely, the autophagy level was lower in the breast cancer tissues. Correlation analysis showed there was a negative correlation between IBP and LC3-II expressions, but significant positive correlation between IBP expression and p-mTOR on Ser2481. These findings for the first time reveal that the mTORC2/Akt/FOXO3a signaling pathway is consistently activated in breast cancer tissues with IBP overexpression. The value of p-mTOR on Ser2481 as a novel high-contrast immunohistochemical mitosis marker in larger populations of human breast carcinomas has been reported.49 These findings support the fact that the activation of the mTORC2/Akt/FOXO3a signaling pathway due to IBP overexpression leads to autophagy suppression, and promotes tumorigenesis in breast cancer.
PI3K is an upstream component of mTORC1 and mTORC2 pathways and its downstream targets include Akt and mTOR. The detail upstream regulators of mTORC2 are still elusive, because mTORC2 includes mTOR, rictor, mLST8 and SIN1.21 So, the detailed effect mechanisms of IBP on mTORC2-related molecules are still not known. It needs to be further elucidated.
Recent work found that autophagy had dual roles of pro-death and pro-survival in different cancers under different environmental and cellular stress.50 In this study, our results showed IBP-mediated inhibition of autophagy represented an important mechanism of tumorigenesis in breast cancer, suggesting that autophagy may serve as a pro-death mechanism in breast cancer development. This seems to be consistent with the other scholars’ reports.14, 36, 37, 38 However, the detailed mechanisms between autophagy and breast cancer growth and metastasis still need to be further elucidated.
IBP has multiple roles in the biological processes of the immune system.51, 52, 53 IBP also had an important role in the progression of malignant tumors such as breast cancer cells, oral squamous cell carcinoma and so on.4, 54, 55 Interestingly, IBP-suppressed cisplatin-mediated apoptosis of breast cancer cells, suggesting IBP may serve as a target for pharmacologic intervention of breast cancer resistant to cisplatin therapy.56 It is known that there are some relationships between autophagy and resistance of cancer cells to anticancer drugs.57 So, it is very interesting to observe the effects of IBP-mediated inhibition autophagy on the resistance of breast cancer cell to anticancer drugs such as cisplatin and paclitaxel and so on.
In summary, IBP-mediated suppression of autophagy promotes breast cancer cell growth by activating the mTORC2/Akt/FOXO3a signaling pathway (Figure 8). The anti-autophagic property of IBP is a key feature of IBP-mediated tumorigenesis, and IBP may, to some extent, serve as an attractive target for breast cancer therapy.
A putative modal for the mechanisms of how suppression of autophagy by IBP promotes breast cancer cell growth and metastasis. IBP could promote breast cancer growth and metastasis by suppressing autophagy through activating the mTORC2/Akt/FOXO3a signaling pathway (measured as the phosphorylation of their substrates: mTOR at Ser2481, Akt at Ser473 and FOXO3a at Thr32)
Materials and Methods
Cell culture
MDA-MB-231 and MDA-MB-468 breast cancer cell lines were purchased from the Cell Bank of the Committee on Type Culture Collection of Chinese Academy of Sciences (CCTCC, Shanghai, China). The MDA-MB-231 cells were cultured in RPMI 1640 medium (Gibco, Carlsbad, CA, USA), containing 100 U/ml penicillin, 0.1 mg/ml streptomycin and 10% fetal bovine serum (FBS). Antibiotics and FBS were purchased from HyClone (Waltham, MA, USA). The MDA-MB-468 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% FBS and antibiotics. The cells were maintained under an atmosphere of 95% air and 5% CO2 at 37 °C. Then appropriate cells were starved using Hank’s balanced salt solution (HBSS; starvation condition) purchased from HyClone.
Reagents
Opti-MEM I, Lipofectamine 2000 reagent and blasticidin were purchased from Invitrogen (Carlsbad, CA, USA). mTOR C2/mTOR C1 inhibitor OSI-027 was purchased from Active Biochemicals Company (Hongkong, China). Rapamycin, thiazolyl blue tetrazolium bromide (MTT), bafilomycin and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich (Louis, MO, USA). PI3K inhibitor LY294002 (LY) (Beyotime, Hangzhou, China), antibodies against microtubule-associated protein 1 light chain 3 (LC3), mTOR, p-mTOR (Ser2448), p-mTOR (Ser2481), Akt, p-Akt (Ser473), Foxo3a, p-Foxo1 (Thr24), Foxo3a (Thr32), p70 S6 kinase, p-p70 S6 kinase (T389), 4EBP1, p-4EBP1 (Thr37/46), p62, were from Cell Signaling Technology (Boston, MA, USA). RFP-LC3 vector was kindly provided by Yoshimori T at Osaka University, Japan.
RFP-LC3 assay
Cells were transfected with RFP-LC3 vector using Lipofectamine 2000. After 48 h, cells were treated with HBSS, then fixed in 4% formaldehyde for 20 min, and observed under a laser scanning confocal microscope (Leica, Heidelberg, Germany).
Transmission electron microscopy (TEM)
Cells (5 × 105) were seeded in 3-cm dishes and allowed to attach overnight. The cells were treated with and without HBSS for 0.5 h at 37°C. Cells and tissues from breast cancer patients were fixed in ice-cold 2.5% glutaraldehyde (in 0.1 mol/L PBS, pH 7.3), post-fixed in 1% OsO4 and 0.1% potassium ferricyanide, dehydrated through a graded series of ethanol (30–90%), and embedded in Epon. Ultrathin sections (300 nm) were obtained using a Reichart Ultracut (Leica Microsystems, Chicago, IL, USA), stained with 0.5% toluidine blue, and examined under light microscope. Ultrathin sections (65 nm) were stained with 2% uranyl acetate and Reynold’s lead citrate and examined using TECNAI 10 transmission electron microscope (Hillsboro, OR, USA).
qRT-PCR analysis
Total RNA was extracted from treated cells using TRIZOL (Invitrogen). Reverse transcription was performed using ReverTra Ace-α-kits (Toyobo, Osaka, Japan). The PCR data were validated by qRT-PCR using SYBR green PCR master mix (Tingen Biotech, Beijing, China) according to the manufacturer’s instructions.
Western blot analysis
Total cell lysates were prepared with RIPA buffer (Beyotime). For Western blot assay, proteins (40 μg/well) were separated by 10% sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA, USA). The membrane was incubated in blocking buffer (Tris-buffered saline (TBS), containing 0.1% Tween 20 and 5% BSA) for 2 h at room temperature. Upon incubation with primary and secondary antibodies, immunoreactive bands were detected with ECL kit (Millipore) according to the manufacturer’s instructions.
Overexpression and RNA interference of IBP in breast cancer cells
MDA-MB-468 cells were individually transfected with 1 μg of pEGFP-C1 IBP expression plasmid or control vector using Lipofectamine 2000 according to the manufacturer's instructions. After 24 h, the transfected cells were split at 1:20. The cell growth medium was supplemented with G418 (800 μg/ml) to select cells with successful transfection 2 days after transfection. For RNA interference, two sequences targeting IBP were used for RNA interference (178: 5′-GCTGAAGCAGTTGATGCAG-3′ and 568: 5′-CAGCAATGAGCAGCAGAAC)-3′. An unrelated sequence was used as a negative control (5′-GTCTCCGAACGTGTCACGT-3′). These sequences were individually subcloned into GeneBuster vector (Genordia, Bromma, Sweden) to obtain recombinant expression plasmids. MDA-MB-231 cells were transfected with either IBP siRNA recombinant plasmid or negative control vector. Growth medium supplemented with Blasticidin S (1 mg/ml) was employed to select cells with successful transfection, which were neomycin resistant.
MTT assay for proliferation
Cells were seeded at 2 × 103 cells/well in 200 μl of medium in triplicate in 96-well plates and cultured for 24 h. Then, cells were treated with or without rapamycin, OSI-027 or both for 48 h. The number of viable cells transfected with either IBP overexpressing or downregulating plasmid was determined by MTT assay. In brief, 20 μl of MTT solution (5 mg/ml in PBS) was added to each well, followed by incubation for 4 h at 37 °C. Then, the solution was removed, and 150 μl of dimethyl sulfoxide was added to each well. These plates were rocked gently for 10 min and the absorbance was measured with a microplate reader (Bio-RAD Model 550, Hercules, CA, USA) at 490 nm.
Wound-healing assay
For wound-healing assay, MDA-MB-231 cells (3 × 105) were seeded in 6-well plates and grown until 100% confluence. The confluent monolayer was wounded with a sterile micropipette tip. Detached cells were removed by washing with PBS and cells were incubated in complete medium with or without rapamycin (10 nM), OSI-027 (10 μM) or both for 48 h. Then, representative photographs were captured under light microscope and the healing was evaluated.
Transwell migration assay
Transwell migration assay was conducted using a Transwell chamber (Millicell, Cat. PI8P01250; Millipore) that is 12 mm in diameter and 8 mm in pore size. The upper chamber was coated with 100 μl of extracellular matrix (ECM) gel (Catalog No. E1270, Sigma) at a final concentration of 1 : 2 at room temperature for 20 min. Breast cancer cells in serum-free medium (5 × 104 cells per well) were added to the upper chamber, and conditional medium with or without rapamycin (10 nM), OSI-027 (10 μM) or both was placed into the lower chamber as a chemoattractant followed by incubation for 48 h at 37 °C with 5% CO2. Migrated cells on the undersides of filter membrane were then fixed in 4% paraformaldehyde and stained with crystal violet (Beyotime). Migrated cells were counted in five randomly selected fields of each well under a microscope.
Colony formation assay
Cells were directly seeded (200 cells/well) into 6-well plates, allowed to attach overnight, and then treated with or without rapamycin (10 nM), OSI-027 (10 μM) or both for 2 weeks. Colonies were fixed and stained with crystal violet. Excess dye was removed by rinsing with tap water, and the plates were air-dried. Colonies were counted under a microscope.
In vivo tumorigenicity assay
For tumorigenicity assay, 4-week-old BALB/c athymic nude mice weighing 20–25 g (Experimental Animal Center, The Third Military Medical University, Chongqing, China) were subcutaneously injected with 9 × 106 IBP-siRNA-MDA-MB-231 cells or control-siRNA-MDA-MB-231 cells into the flanks on the right side of each mouse. Four days later, the control siRNA group was divided into four groups (n=7). Mice of each group were intravenously injected with rapamycin (8 mg/kg), OSI-027 (10 mg/kg), rapamycin+OSI-027 and DMSO, respectively. Tumor length and width were measured every third day with a caliper and the tumor volumes were calculated from the equation (length × width2)/2. Mice were killed thirty days later. Tumors and organs were collected from mice, fixed in 4% formaldehyde, sectioned and routinely stained with hematoxylin and eosin (H&E). The lung and liver micrometastatic foci were observed under inverted microscope (NIKON, E100, Tokyo, Japan).
Tissue array
A human breast cancer tissue array (BC081116a) was purchased from the Baisida Biotechnology (Xi’an, China) and detection was conducted according to the manufacturer’s instructions. Protein expression was scored semi-quantitatively based on the staining intensity (SI) and distribution using the immunoreactive score (IRS). Briefly, IRS=SI × PP (percentage of positive cells). SI was determined as 0=negative; 1=weak; 2=moderate; and 3=strong. PP was defined as 1, <25%; 2, 25–50%; 3, 50–75%; and 4, 75–100% positive cells. IRS ≥4 was defined as positive.
Sample collection
Breast cancer tissues and adjacent normal tissues were collected from seven patients undergoing surgical intervention due to primary breast cancer in the Daping Hospital of Third Military Medical University. None underwent chemotherapy or radiotherapy before surgery. Tissues were sectioned and stained with H&E. The study protocols were approved by the local Institutional Review Board (IRB), and informed consent was obtained from patients.
Immunohistochemistry assay
Immunohistochemistry staining was carried out with an indirect immunoperoxidase method using antibodies against IBP (1 : 200), p-mTOR (Ser2481; 1 : 50), LC3 (1 : 200), p-Akt (Ser473, 1 : 50), and p-FOXO3a (Thr32; 1 : 50).
Statistical analysis
The statistical significance of differences between groups was assessed using the SPSS (SPSS Inc., Chicago, IL, USA). Data were assessed using unpaired two-tailed t-test and regression analysis. A value of P<0.05 was considered statistically significant.
Abbreviations
- RFP:
-
red fluorescent protein
- IBP:
-
interferon regulatory factor-4 binding protein
- qRT-PCR:
-
quantitative real-time PCR
- FBS:
-
fetal bovine serum
- PI3K:
-
phosphatidylinositol 3-kinase
- WT:
-
wild type
- mTOR:
-
mammalian target of rapamycin
- mTORC1:
-
mammalian target of rapamycin complex 1
- mTORC2:
-
mammalian target of rapamycin complex 2
- MTT:
-
methyl thiazolyl tetrazolium
- HBSS:
-
Hank's Balanced Salt Solution
- p70S6K:
-
p70 ribosomal protein S6 kinase
- Akt (PKB):
-
protein kinase B
- TEM:
-
transmission electron microscopy
- FOXO3a:
-
Forkhead family of transcription factors 3a
- 4EBP1:
-
eukaryotic translation initiation factor 4E (eIF-4E) binding protein
- LC3:
-
microtubule-associated protein 1 light chain 3
References
Allan AL, Vantyghem SA, Tuck AB, Chambers AF . Tumor dormancy and cancer stem cells: implications for the biology and treatment of breast cancer metastasis. Breast Dis 2006; 26: 87–98.
O'Shaughnessy J . Extending survival with chemotherapy in metastatic breast cancer. Oncologist 2005; 10 (Suppl 3): 20–29.
Tanaka Y, Bi K, Kitamura R, Hong S, Altman Y, Matsumoto A et al. SWAP-70-like adapter of T cells, an adapter protein that regulates early TCR-initiated signaling in Th2 lineage cells. Immunity 2003; 18: 403–414.
Li P, Zhang Z, Wang Q, Li S, Zhang Y, Bian X et al. The ectopic expression of IFN regulatory factor 4-binding protein is correlated with the malignant behavior of human breast cancer cells. Int Immunopharmacol 2009; 9: 1002–1009.
Hoyer-Hansen M, Jaattela M . Autophagy: an emerging target for cancer therapy. Autophagy 2008; 4: 574–580.
Baehrecke EH . Autophagy: dual roles in life and death? Nat Rev Mol Cell Biol 2005; 6: 505–510.
Edinger AL, Thompson CB . Defective autophagy leads to cancer. Cancer Cell 2003; 4: 422–424.
Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999; 402: 672–676.
Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 2003; 112: 1809–1820.
Levine B . Cell biology: autophagy and cancer. Nature 2007; 446: 745–747.
Amaravadi RK, Thompson CB . The roles of therapy-induced autophagy and necrosis in cancer treatment. Clin Cancer Res 2007; 13: 7271–7279.
Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev 2007; 21: 1621–1635.
Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell 2009; 137: 1062–1075.
Oh S, Xiaofei E, Ni D, Pirooz SD, Lee JY, Lee D et al. Downregulation of autophagy by Bcl-2 promotes MCF7 breast cancer cell growth independent of its inhibition of apoptosis. Cell Death Differ 2011; 18: 452–464.
Mavrakis KJ, McKinlay KJ, Jones P, Sablitzky F . DEF6 a novel PH-DH-like domain protein, is an upstream activator of the Rho GTPases Rac1, Cdc42, and RhoA. Exp Cell Res 2004; 294: 335–344.
LoPiccolo J, Blumenthal GM, Bernstein WB, Dennis PA . Targeting the PI3K/Akt/mTOR pathway: effective combinations and clinical considerations. Drug Resist Updat 2008; 11: 32–50.
Yap TA, Garrett MD, Walton MI, Raynaud F, de Bono JS, Workman P . Targeting the PI3K-AKT-mTOR pathway: progress, pitfalls, and promises. Curr Opin Pharmacol 2008; 8: 393–412.
Bjornsti MA, Houghton PJ . The TOR pathway: a target for cancer therapy. Nat Rev Cancer 2004; 4: 335–348.
Hay N . The Akt-mTOR tango and its relevance to cancer. Cancer Cell 2005; 8: 179–183.
Guertin DA, Sabatini DM . Defining the role of mTOR in cancer. Cancer Cell 2007; 12: 9–22.
Bhaskar PT, Hay N . The two TORCs and Akt. Dev Cell 2007; 12: 487–502.
Chen CH, Sarbassov dos D . The mTOR (mammalian target of rapamycin) kinase maintains integrity of mTOR complex 2. J Biol Chem 2011; 286: 40386–40394.
Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell 2006; 11: 859–871.
Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab 2007; 6: 458–471.
Mizushima N, Yoshimori T, Levine B . Methods in mammalian autophagy research. Cell 2010; 140: 313–326.
Klionsky DJ, Cuervo AM, Seglen PO . Methods for monitoring autophagy from yeast to human. Autophagy 2007; 3: 181–206.
Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 2005; 171: 603–614.
Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282: 24131–24145.
Klionsky DJ, Elazar Z, Seglen PO, Rubinsztein DC . Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy 2008; 4: 849–950.
Liu JJ, Lin M, Yu JY, Liu B, Bao JK . Targeting apoptotic and autophagic pathways for cancer therapeutics. Cancer Lett 2011; 300: 105–114.
Copp J, Manning G, Hunter T . TORC-specific phosphorylation of mammalian target of rapamycin (mTOR): phospho-Ser2481 is a marker for intact mTOR signaling complex 2. Cancer Res 2009; 69: 1821–1827.
Inoki K, Li Y, Xu T, Guan KL . Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev 2003; 17: 1829–1834.
Inoki K, Li Y, Zhu T, Wu J, Guan KL . TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol 2002; 4: 648–657.
Tu YF, Kaipparettu BA, Ma Y, Wong LJ . Mitochondria of highly metastatic breast cancer cell line MDA-MB-231 exhibits increased autophagic properties. Biochim Biophys Acta 2011; 1807: 1125–1132.
Mizushima N, Levine B, Cuervo AM, Klionsky DJ . Autophagy fights disease through cellular self-digestion. Nature 2008; 451: 1069–1075.
Rubinsztein DC . The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 2006; 443: 780–786.
Akar U, Chaves-Reyez A, Barria M, Tari A, Sanguino A, Kondo Y et al. Silencing of Bcl-2 expression by small interfering RNA induces autophagic cell death in MCF-7 breast cancer cells. Autophagy 2008; 4: 669–679.
Indelicato M, Pucci B, Schito L, Reali V, Aventaggiato M, Mazzarino MC et al. Role of hypoxia and autophagy in MDA-MB-231 invasiveness. J Cell Physiol 2010; 223: 359–368.
Chiavarina B, Whitaker-Menezes D, Migneco G, Martinez-Outschoorn UE, Pavlides S, Howell A et al. HIF1-alpha functions as a tumor promoter in cancer associated fibroblasts, and as a tumor suppressor in breast cancer cells: Autophagy drives compartment-specific oncogenesis. Cell Cycle 2010; 9: 3534–3551.
Chapuis N, Tamburini J, Green AS, Willems L, Bardet V, Park S et al. Perspectives on inhibiting mTOR as a future treatment strategy for hematological malignancies. Leukemia 2010; 24: 1686–1699.
Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002; 110: 163–175.
Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol 2004; 14: 1296–1302.
Carayol N, Vakana E, Sassano A, Kaur S, Goussetis DJ, Glaser H et al. Critical roles for mTORC2- and rapamycin-insensitive mTORC1-complexes in growth and survival of BCR-ABL-expressing leukemic cells. Proc Natl Acad Sci USA 2010; 107: 12469–12474.
Sarbassov DD, Guertin DA, Ali SM, Sabatini DM . Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 2005; 307: 1098–1101.
Hresko RC, Mueckler M . mTOR.RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J Biol Chem 2005; 280: 40406–40416.
Hay N, Sonenberg N . Upstream and downstream of mTOR. Genes Dev 2004; 18: 1926–1945.
Richter JD, Sonenberg N . Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 2005; 433: 477–480.
Bhagwat SV, Gokhale PC, Crew AP, Cooke A, Yao Y, Mantis C et al. Preclinical characterization of OSI-027, a potent and selective inhibitor of mTORC1 and mTORC2: distinct from rapamycin. Mol Cancer Ther 2011; 10: 1394–1406.
Lopez-Bonet E, Vazquez-Martin A, Perez-Martinez MC, Oliveras-Ferraros C, Perez-Bueno F, Bernado L et al. Serine 2481-autophosphorylation of mammalian target of rapamycin (mTOR) couples with chromosome condensation and segregation during mitosis: confocal microscopy characterization and immunohistochemical validation of PP-mTOR(Ser2481) as a novel high-contrast mitosis marker in breast cancer core biopsies. Int J Oncol 2010; 36: 107–115.
Dalby KN, Tekedereli I, Lopez-Berestein G, Ozpolat B . Targeting the prodeath and prosurvival functions of autophagy as novel therapeutic strategies in cancer. Autophagy 2010; 6: 322–329.
Becart S, Charvet C, Canonigo Balancio AJ, De Trez C, Tanaka Y, Duan W et al. SLAT regulates Th1 and Th2 inflammatory responses by controlling Ca2+/NFAT signaling. J Clin Invest 2007; 117: 2164–2175.
Canonigo-Balancio AJ, Fos C, Prod'homme T, Becart S, Altman A . SLAT/Def6 plays a critical role in the development of Th17 cell-mediated experimental autoimmune encephalomyelitis. J Immunol 2009; 183: 7259–7267.
Gupta S, Lee A, Hu C, Fanzo J, Goldberg I, Cattoretti G et al. Molecular cloning of IBP, a SWAP-70 homologous GEF, which is highly expressed in the immune system. Hum Immunol 2003; 64: 389–401.
Zhang Z, Wang Q, Li P, Zhou Y, Li S, Yi W et al. Overexpression of the Interferon regulatory factor 4-binding protein in human colorectal cancer and its clinical significance. Cancer Epidemiol 2009; 33: 130–136.
Jian CX, Yang MZ, Li P, Xiong J, Zhang ZJ, Li CJ et al. Ectopically expressed IBP promotes cell proliferation in oral squamous cell carcinoma. Cancer Invest 2012; 30: 748–756.
Yang M, Yuan F, Li P, Chen Z, Chen A, Li S et al. Interferon regulatory factor 4 binding protein is a novel p53 target gene and suppresses cisplatin-induced apoptosis of breast cancer cells. Mol Cancer 2012; 11: 54.
Aredia F, Scovassi AI . Manipulation of autophagy in cancer cells: an innovative strategy to fight drug resistance. Future Med Chem 2013; 5: 1009–1021.
Acknowledgements
We thank Dr. Yoshimori (Osaka University, Japan) for providing mRFP-LC3 plasmid and Dr. Jiqing Lian for providing good advice. This project was supported by the National Natural Science Foundation of China (81272909 and 81072154).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by GM Fimia
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Chen, S., Han, Q., Wang, X. et al. IBP-mediated suppression of autophagy promotes growth and metastasis of breast cancer cells via activating mTORC2/Akt/FOXO3a signaling pathway. Cell Death Dis 4, e842 (2013). https://doi.org/10.1038/cddis.2013.380
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.380
Keywords
This article is cited by
-
TSSC3 promotes autophagy via inactivating the Src-mediated PI3K/Akt/mTOR pathway to suppress tumorigenesis and metastasis in osteosarcoma, and predicts a favorable prognosis
Journal of Experimental & Clinical Cancer Research (2018)
-
O-GlcNAcylation mediates metastasis of cholangiocarcinoma through FOXO3 and MAN1A1
Oncogene (2018)
-
Downregulation of ATG5-dependent macroautophagy by chaperone-mediated autophagy promotes breast cancer cell metastasis
Scientific Reports (2017)
-
Deconvoluting the relationships between autophagy and metastasis for potential cancer therapy
Apoptosis (2016)
-
Nifuroxazide induces apoptosis and impairs pulmonary metastasis in breast cancer model
Cell Death & Disease (2015)