Abstract
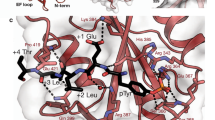
Cbl is an adaptor protein that functions as a negative regulator of many signalling pathways that start from receptors at the cell surface1,2,3,4. The evolutionarily conserved amino-terminal region of Cbl (Cbl-N) binds to phosphorylated tyrosine residues and has cell-transforming activity. Point mutations in Cbl that disrupt its recognition of phosphotyrosine also interfere with its negative regulatory function and, in the case of v-cbl, with its oncogenic potential5. In T cells, Cbl-N binds to the tyrosine-phosphorylated inhibitory site of the protein tyrosine kinase ZAP-706. Here we describe the crystal structure of Cbl-N, both alone and in complex with a phosphopeptide that represents its binding site in ZAP-70. The structures show that Cbl-N is composed of three interacting domains: a four-helix bundle (4H), an EF-hand7 calcium-binding domain, and a divergent SH2 domain8 that was not recognizable from the amino-acid sequence of the protein. The calcium-bound EF hand wedges between the 4H and SH2 domains and roughly determines their relative orientation. In the ligand-occupied structure, the 4H domain packs against the SH2 domain and completes its phosphotyrosine-recognition pocket. Disruption of this binding to ZAP-70 as a result of structure-based mutations in the 4H, EF-hand and SH2 domains confirms that the three domains together form an integrated phosphoprotein-recognition module.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Thien, C. B. & Langdon, W. Y. c-Cbl: a regulator of T cell receptor-mediated signalling. Immunol. Cell Biol. 76, 473–482 (1998).
Liu, Y. C. & Altman, A. Cbl: Complex formation and functional implications. Cell Signal. 10, 377–385 (1998).
Miyake, S.et al. The Cbl proto-oncogene product: From enigmatic oncogene to center stage of signal transduction. Crit. Rev. Oncogen. 8, 189–218 (1998).
Wange, R. L. & Samelson, L. E. Complex complexes: signaling at the TCR. Immunity 5, 197–205 (1996).
Thien, C. B. & Langdon, W. Y. EGF receptor binding and transformation by v-cbl is ablated by the introduction of a loss-of-function mutation from the Caenorhabditis elegans sli-1 gene. Oncogene 14, 2239–2249 (1997).
Lupher, M. L. J, Songyang, Z., Shoelson, S. E., Cantley, L. C. & Band, H. The Cbl phosphotyrosine-binding domain selects a D(N/D)XpY motif and binds to the Tyr292 negative regulatory phosphorylation site of ZAP-70. J. Biol. Chem. 272, 33140–33144 (1997).
Ikura, M. Calcium binding and conformational response in EF-hand proteins. Trends Biochem. Sci. 21, 14–17 (1996).
Kuriyan, J. & Cowburn, D. Modular peptide recognition domains in eukaryotic signaling. Annu. Rev. Biophys. Biomol. Struct. 26, 259–288 (1997).
Saurin, A. J., Borden, K. L., Boddy, M. N. & Freemont, P. S. Does this have a familiar RING? Trends Biochem. Sci. 21, 208–214 (1996).
Blake, T. J., Shapiro, M., Morse, H. C. & Langdon, W. Y. The sequences of the human and mouse c-cbl proto-oncogenes show v-cbl was generated by a large truncation encompassing a proline-rich domain and a leucine zipper-like motif. Oncogene 6, 653–657 (1991).
Galisteo, M. L., Dikic, I., Batzer, A. G., Langdon, W. Y. & Schlessinger, J. Tyrosine phosphorylation of the c-cbl proto-oncogene protein product and association with epidermal growth factor (EGF) receptor upon EGF stimulation. J. Biol. Chem. 270, 20242–20245 (1995).
Deckert, M., Elly, C., Altman, A. & Liu, Y. C. Coordinated regulation of the tyrosine phosphorylation of Cbl by Fyn and Syk tyrosine kinases. J. Biol. Chem. 273, 8867–8874 (1998).
Yoon, C. H., Lee, J., Jongeward, G. D. & Sternberg, P. W. Similarity of sli-1, a regulator of vulval development in C. elegans, to the mammalian proto-oncogene c-cbl. Science 269, 1102–1105 (1995).
Meisner, H. etal. Interactions of Drosophila Cbl with epidermal growth factor receptors and role of Cbl in R7 photoreceptor cell development. Mol. Cell Biol. 17, 2217–2225 (1997).
Ota, Y. & Samelson, L. E. The product of the proto-oncogene c-cbl: a negative regulator of the Syk tyrosine kinase. Science 276, 418–420 (1997).
Holm, L. & Sander, C. Dali anetwork tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 (1995).
Kretsinger, R. H. EF-hands embrace. Nature Struct. Biol. 4, 514–516 (1997).
Essen, L. O., Perisic, O., Cheung, R., Katan, M. & Williams, R. L. Crystal structure of a mammalian phosphoinositide-specific phospholipase Cδ. Nature 380, 595–602 (1996).
de Beer, T., Carter, R. E., Lobel-Rice, K. E., Sorkin, A. & Overduin, M. Structure and Asn-Pro-Phe binding pocket of the Eps15 homology domain. Science 281, 1357–1360 (1998).
Becker, S., Groner, B. & Muller, C. W. Three-dimensional structure of the Stat3β homodimer bound to DNA. Nature 394, 145–151 (1998).
Chen, X.et al. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell 93, 827–839 (1998).
Hatada, M. H. etal. Molecular basis for the interaction of the protein tyrosine kinase ZAP-70 with the T-cell receptor. Nature 377, 32–38 (1995).
Eck, M. J., Shoelson, S. E. & Harrison, S. C. Recognition of a high-affinity phosphotyrosyl peptide by the Src homology-2 domain of p56lck. Nature 362, 87–91 (1993).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Collaborative Computational Project Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D 50, 760–776 (1994).
Jones, T. A., Zhou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brunger, A. X-PLOR Version 3.0: A System for Crystallography and NMR (Yale University Press, New Haven, (1992).
Navaza, J. (ed. AMoRe: A New Package for Molecular Replacement (SERC, Daresbury, UK, (1992).
Kraulis, P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Nicholls, A., Sharp, K. A. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 11, 281–296 (1991).
Acknowledgements
We thank C. Dahl for synthesis and purification of the ZAP-70 phosphopeptide, the staff at MacCHESS for assistance with data collection, and S. Harrison and T. Roberts for comments on the manuscript. M.J.E. is a recipient of a Burroughs–Wellcome Career Award in the Biomedical Sciences. Diffraction data were recorded at the Cornell High Energy Synchrotron Source (CHESS), which is supported by grants from the NSF and NIH.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Meng, W., Sawasdikosol, S., Burakoff, S. et al. Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase. Nature 398, 84–90 (1999). https://doi.org/10.1038/18050
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/18050
This article is cited by
-
The co-crystal structure of Cbl-b and a small-molecule inhibitor reveals the mechanism of Cbl-b inhibition
Communications Biology (2023)
-
Cbl-mediated K63-linked ubiquitination of JAK2 enhances JAK2 phosphorylation and signal transduction
Scientific Reports (2017)
-
Ubiquitination switches EphA2 vesicular traffic from a continuous safeguard to a finite signalling mode
Nature Communications (2015)
-
A PKC-SHP1 signaling axis desensitizes Fcγ receptor signaling by reducing the tyrosine phosphorylation of CBL and regulates FcγR mediated phagocytosis
BMC Immunology (2014)
-
JAK–cytokine receptor recognition, unboxed
Nature Structural & Molecular Biology (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.