Abstract
Exudative age-related macular degeneration (AMD) and polypoidal choroidal vasculopathy (PCV) share similar abnormal choroidal vasculature, but responses to treatments are different. In this study, we sequenced the whole HTRA1 gene and its promoter by direct sequencing in a Hong Kong Chinese PCV cohort. We identified rs11200638, c.34delCinsTCCT, c.59C>T, rs1049331 and rs2293870 significantly associated with PCV. Notably, rs2672598 was significantly associated with exudative AMD (p = 1.31 × 10−4) than PCV (p = 0.11). Logistic regression indicated that rs2672598 (p = 2.27 × 10−3) remained significant after adjusting for rs11200638 in exudative AMD. Moreover, the rs11200638-rs2672598 joint genotype AA-CC conferred higher risk to exudative AMD (43.11 folds) than PCV (3.68 folds). Promoter analysis showed that rs2672598 C-allele showed higher luciferase expression than wildtype T-allele (p = 0.026), independent of rs11200638 genotype (p = 0.621). Coherently, vitreous humor HTRA1 expression with rs2672598 CC genotype was significantly higher than that with TT genotype by 2.56 folds (p = 0.02). Furthermore, rs2672598 C-allele was predicted to alter the transcription factor binding sites, but not rs11200638 A-allele. Our results revealed that HTRA1 rs2672598 is more significantly associated with exudative AMD than PCV in ARMS2/HTRA1 region, and it is responsible for elevated HTRA1 transcriptional activity and HTRA1 protein expression.
Similar content being viewed by others
Introduction
Age-related macular degeneration (AMD) is a progressive macular disease and a major cause of irreversible blindness in the elderly population1. At the advanced stage, it can be classified into exudative and non-exudative AMD2. Exudative AMD, characterized by choroidal neovascularization (CNV), shares similar phenotype with polypoidal choroidal vasculopathy (PCV). Both of them involve abnormal vasculopathies arising from the choroidal vasculature leading to sub-retinal serous exudation and hemorrhage3,4. PCV is characterized by the polypoidal structures beneath the relatively intact Bruch’s membrane, branching from abnormal inner choroidal vasculature5. The polypoidal structures can be visualized by indocyanine green angiography (ICGA), a diagnostic tool for differentiating PCV lesion from CNV. PCV and CNV are different in their risk factors6, clinical manifestations7, natural courses5, responses to treatment8, and the overall visual prognosis9. PCV occurs predominately in Asian populations and tends to be younger6. Its disease course is relatively benign with less drusen but a tendency of relapse5. Without treatment, the visual outcome of PCV is usually guarded10. Patients with AMD, however, tend to present with a more aggressive course and uncontrollable visual deterioration6. In addition, exudative AMD responds better to anti-vascular endothelial growth factor (VEGF) treatment, whereas PCV better to photodynamic therapy (PDT)9,11,12.
Some reports propose that PCV could be a variant of typical CNV and regarded as one of the subtypes of exudative AMD6,13. This could be reflected by their similar genetic and environmental factors. Multiple genetic variants have exhibited significant association with both exudative AMD and PCV6. These include the complement factor H (CFH) gene, age-related maculopathy susceptibility 2 (ARMS2)/high temperature requirement factor A1 (HTRA1) locus and complement component 2 (C2)/factor B (CFB) genes14. Previously, we screened the entire ARMS2 gene by direct sequencing in our Hong Kong Chinese exudative AMD and PCV cohort15. Although significant differences in the genotypic distributions of 11 polymorphisms in ARMS2 gene were found between exudative AMD and PCV, all of the associated polymorphisms showed similar trends. Since the ARMS2 and HTRA1 genes are in complete linkage disequilibrium (LD), it is also important to delineate the association pattern of HTRA1 in PCV patients. Three HTRA1 haplotypes are associated individually with typical AMD and PCV in a Japanese study although no difference between them was reported16. Therefore, the differential association between exudative AMD and PCV remains equivocal. The association of other HTRA1 variants in PCV should be investigated. In this study, 9 exons, intron-exon junctions and the promoter region of HTRA1 gene were sequenced in our Hong Kong Chinese PCV cohort with a view to clarify the association of HTRA1 with PCV.
Results
Sequence variant association analysis
Totally 26 polymorphisms in HTRA1 were identified in PCV patients (Table 1). Four of them, IVS5+76_79delGTTT, rs79778361 (IVS6+111G>A), rs76357476 (IVS7+149C>G) and rs2293871 (IVS8-36C>T), were excluded from further analysis since they did not follow the Hardy-Weinberg equilibrium. Among the remaining 22 polymorphisms, 10 uncommon variants (minor allele frequency <3%) and 6 common variants did not showed significant associations. They were excluded in subsequent analysis. The remaining 6 variants were further analyzed for their corresponding ORs under dominant model (Table 2). Two SNPs, c.34delCinsTCCT and rs2672598 (−487T>C), were also included in the analysis since they showed significant association with exudative AMD. Four variants were associated with exudative AMD: rs11200638 (−625G>A; p = 1.16 × 10−5, OR = 3.21, 95% confident intervals (CI): 1.88–5.50), c.34delCinsTCCT (L12insS; p = 0.001, OR = 0.13, 95% CI: 0.13–0.57), rs1049331 (c.102C>T, A34A; p = 1.16 × 10−5, OR = 3.21, 95% CI: 1.88–5.50), and rs2293870 (c.108G>T, G36G; p = 1.16 × 10−5, OR = 3.21, 95% CI: 1.88–5.50) as well as PCV (rs11200638: p = 0.005, OR = 2.01, 95% CI: 1.23–3.28; c.34delCinsTCCT: p = 0.029, OR = 0.34, 95% CI: 0.13–0.90; rs1049331: p = 0.009, OR = 1.91, 95% CI: 1.17–3.11; and rs2293870: p = 0.009, OR = 1.91, 95% CI: 1.17–3.11). None of these variants showed significant differences between AMD and PCV under dominant model (p > 0.05). Instead, a significant differential association (p = 0.015, OR = 5.48, 95% CI: 1.19–25.37) was detected in rs2672598. It was strongly associated with exudative AMD (p = 1.31 × 10−4, OR = 10.53, 95% CI: 2.41–46.04) but not associated with PCV (p = 0.106). Our result is coherent to a recent report in northern China that rs2672598 is associated with exudative AMD (p = 4.60 × 10−3), but not PCV (p = 0.24)17. In order to further illustrate the association of rs2672598 with PCV, we genotyped another PCV cohort in southern China. Unlike the Hong Kong and Beijing cohort, rs2672598 was associated with PCV in Shantou cohort (p = 0.031, OR = 2.49, 95% CI: 1.06–5.83). The meta-analysis of the 3 Chinese cohorts at rs2672598 also showed a significant association with PCV (p = 0.02, OR = 1.94, 95% CI: 1.13–3.32; Fig. 1). In addition, c.59C>T (A20V) was significantly associated with PCV (p = 0.004, OR = 0.49, 95% CI: 0.30–0.79), but not with exudative AMD (p = 0.13).
Three Chinese cohorts (Hong Kong, Shantou and Beijing) were included in the meta-analysis under dominant model. Random effect was chosen. Squares indicate study-specific OR. The size of the box is proportional to the weight of the study. Horizontal lines indicate 95% CI. Diamond indicates summary OR with its corresponding 95% CI.
Haplotype analysis
LD and haplotype-based analyses of the 6 associated variants showed that rs11200638, rs1049331, and rs2293870 were in perfect LD (r2 > 0.95) in both exudative AMD and PCV. LD blocks across all variants were observed in exudative AMD (Fig. 2A), PCV (Fig. 2B) and in control subjects (Fig. 2C), except c.34delCinsTCCT probably because of its low frequency. It was excluded for haplotype-based association analysis. Three associated haplotypes defined by these 5 variants were predicted (Table 3). Haplotype ACCTT, defined by the risk alleles, was associated with increasing risk in both exudative AMD (p = 8.70 × 10−14, OR = 3.10, 95% CI: 2.29–4.19) and PCV (p = 1.60 × 10−4, OR = 1.66, 95% CI: 1.24–2.22). Haplotype GTCCG, defined by the wildtype alleles, was associated with decreasing risk in exudative AMD (p = 5.35 × 10−12, OR = 0.25, 95% CI: 0.16–0.38), but not in PCV (p = 0.17). These 2 haplotypes were different between AMD and PCV (p < 0.001). Another haplotype, GCTCG, was associated with PCV (p = 0.04, OR = 0.57, 95% CI: 0.37–0.89), but not with exudative AMD (p = 0.363).
The haplotype analysis revealed linkage disequilibrium (LD) blocks lying across all of the associated HTRA1 polymorphisms except for the c.34delCinsTCCT in (A) exudative AMD, in (B) PCV and in (C) control subjects. The displayed value of the LD was D’ (the value of D’ × 100, threshold = 0.8), and the r2 threshold was 0.8.
Joint genotype analysis
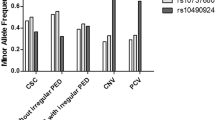
The 4 associated variants between exudative AMD and PCV were examined by joint genotype analysis SNP rs11200638 was used as a proxy because it was in perfect LD with other two SNPs (rs1049331 and rs2293870). Therefore, only rs11200638 and rs2672598 were included in the analysis. The results of the joint genotypes included homozygous carriers of rs2672598-C risk allele. All genotypes of rs11200638 were associated with exudative AMD (pAA-CC = 4.31 × 10−8, pGA-CC = 3.00 × 10−3 and pGG-CC = 2.00 × 10−3, respectively). Significant differences were found between exudative AMD and PCV (pAA-CC = 4.00 × 10−3, OR = 11.70, 95% CI: 1.45–94.53; pGA-CC = 0.01, OR = 10.16, 95% CI: 1.22–84.81 and pGG-CC = 0.02, OR = 11.25, 95% CI: 1.17–108.41, respectively). However, only the homozygous risk allele carriers were associated with PCV (p = 3.00 × 10−3). The highest OR in homozygous risk allele joint genotypes for exudative AMD (OR = 43.11, 95% CI: 5.56–334.49) has 11.7-fold higher risk (p = 4.00 × 10−3) than for PCV (OR = 3.68, 95% CI: 1.51–8.99) (Fig. 3 and Supplementary Table 1). Logistic regression analysis showed that rs11200638 (p = 1.48 × 10−4) and rs2672598 (p = 2.27 × 10−3) were independently associated with exudative AMD after adjusting for age and gender. On the contrary, in PCV, rs11200638 remained significant (p = 9.05 × 10−3) but not rs2672598 (p = 0.20).
HTRA1 promoter analysis
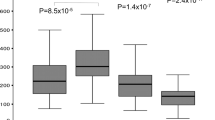
Cloning the HTRA1 promoter identified three rs11200638-rs2672598 haplotypes: G-T (wildtype), G-C and A-C. No A-T haplotype was found. Luciferase expression analysis showed that the rs11200638-rs2672598 haplotypes affect differential transcriptional activities of the HTRA1 promoter (Fig. 4). Comparing to G-T haplotype, elevated transcriptional activity was observed in G-C and A-C haplotypes (p = 0.009, one-way ANOVA; Post-hoc tests adjusted by Tukey HSD: G-C vs G-T, p = 0.026, and A-C vs G-T, p = 0.009) with 1.78 ± 0.35 and 1.99 ± 0.30 fold increase, respectively. However, there was no difference in luciferase expression level between G-C and A-C haplotypes (p = 0.621), suggesting that A-allele of rs11200638 might not alter the transcriptional activity of the HTRA1 promoter. Therefore, rs2672598 should be the variant that contributed to the altered transcriptional activity of the HTRA1 promoter.
A 726-bp genomic fragment (from −658 to +68) containing the HTRA1 promoter was cloned into empty pGL3-Basic vector. Human retinal pigment epithelial cell line ARPE-19 were transfected with different luciferase constructs for 72 hours. (A) Detection of luciferase expression was performed by immunoblotting. (B) Normalized luciferase intensities were calculated by dividing the quantified luciferase intensities by the housekeeping β-actin intensities. Two different clones of each haplotype were used in the transfection experiment. The wildtype rs11200638-rs2672598 haplotype (G-T) was set as a reference. Recombinant firefly luciferase was used as a positive control, whereas empty pGL3 was used as negative control.
Our luciferase-reporter analysis showed that transcription activity increased with rs2672598 C-allele but not rs11200638 A-allele. This phenomenon may be due to influences of transcription factor binding sites within these regions. With G-T haplotype as a reference, one transcription factor E2F-1 binding site was predicted at rs11200638, and one each of STAT4, NFκB, c-Ets-1, RelA, Elk-1 and WT1 binding sites were predicted at rs2672598 site (Fig. 5). When rs11200638 G-allele was replaced by A-allele, the transcription factor E2F-1 binding site was unchanged. In contrast, when rs2672598 T-allele was replaced by C-allele, all the transcription factor binding sites were abolished and changed to GR-alpha, AP-2αA and Sp1 binding sites. Bioinformatic analysis confirmed that rs2672598 contributed to the altered transcriptional activity of the HTRA1 promoter.
DNA sequence of the cloned HTRA1 promoter was used to predict the transcription factor binding sites. PROMO was used to predict the transcription factors that would bind to the region of rs11200638 and rs2672598 in the HTRA1 promoter. Predictions with different alleles of rs11200638 and rs2672598 were performed. Transcription factors: 10: GR-alpha; 12: E2F-1; 13: AP-2αA; 22: STAT4; 23: NFκB; 24: c-Ets-1; 25: RelA; 26: Elk-1; 38: Sp1; 39: WT1.
Correlation of rs2672598 genotype and HTRA1 expression in vitreous humor
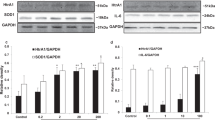
In order to validate the elevated transcriptional activity of rs2672598 C-allele, we performed the genotype-expression correlation analysis in vitreous humor collected from our previous study18, which HTRA1 protein expression has been determined. In this study, the rs2672598 genotype of the vitreous humor samples was determined, and 5 samples were in TT genotype, 23 samples in TC genotype and 27 samples in CC genotypes. The HTRA1 expression was higher in the vitreous humor with rs2672598 CC genotype than that with TT genotype by 2.56 folds (p = 0.009, one-way ANOVA; Post-hoc tests adjusted by Tukey HSD: CC vs TT, p = 0.02; Fig. 6). HTRA1 level of TC genotype is also higher than TT genotype by 2.03 folds but not statistically significant (p = 0.32). As we have previously shown than rs11200638 is not correlated with HTRA1 expression in vitreous humor18, this result confirmed that rs2672598 should be the variant leading to the differential HTRA1 expression in human ocular samples.
HTRA1 expressions in 55 vitreous humor samples were determined by immunoblotting, and expressed in intensity (count/mm2). Genotyping of rs2672598 polymorphism revealed TT genotype in 5 samples, TC genotype in 23 samples and CC genotypes in 27 samples. The HTRA1 expression of CC genotype (homozygous) was significantly higher than that of TT genotype (wildtype).
Discussion
PCV has been regarded as a different disease entity from AMD, with a distinct set of demographic, pathological and clinical characteristics3. However, emerging evidences suggest that PCV could be a variant of typical CNV5, sharing common genetic determinants and environmental risk factors6. Moreover, elevated serum C reactive protein and VEGF levels in CNV were also found in PCV19,20. In clinical practice, PCV usually responds better to PDT8,11, whereas exudative AMD responds better to anti-VEGF treatment12,21. Meanwhile, they share some common genetic determinants. ARMS2 rs10490924 and HTRA1 rs11200638 were associated with both exudative AMD and PCV in a Japanese study, in which neither allelic nor genotypic frequencies showed significant difference22. Similar association of rs10490924 with advanced AMD and PCV has also been reported in Caucasians23. In contrast, our previous study identified significantly different association of ARMS2 polymorphisms between exudative AMD and PCV in the Chinese population15.
Results in this study revealed a HTRA1 promoter variant (rs2672598) that is significantly associated with exudative AMD, but not PCV (Tables 1 and 2). Our result is coherent to a recent report in a northern Chinese population17. Although meta-analysis of the 3 Chinese cohorts at rs2672598 also showed a significant association with PCV (p = 0.02, OR = 1.94, 95% CI: 1.13–3.32; Fig. 1), rs2672598 is more significantly associated with exudative AMD than PCV as shown in Hong Kong (exudative AMD: p = 1.31 × 10−4; PCV: p = 0.11) and Beijing cohorts (exudative AMD: p = 4.60 × 10−3; PCV: p = 0.24). Furthermore, the joint genotype constructed by homozygous carriers of risk allele (rs2672598-C and rs11200638-A) were prone to exudative AMD (OR = 43.11, 95% CI: 5.56–334.49), conferring 11.7-fold higher risk (p = 4.00 × 10−3) when compared to PCV (OR = 3.68, 95% CI: 1.51–8.99; Fig. 3 and Supplementary Table 1). Moreover, joint genotypes constructed by rs2672598-CC and rs11200638-GA/GG conferred increasing AMD risk (OR = 12.86, 95% CI: 1.64–100.86; OR = 18.00, 95% CI: 2.00–161.83, respectively), but not associated with PCV. Logistic regression results further indicated differential roles of rs2672598 in exudative AMD and PCV. This SNP might contribute independently of rs11200638 to exudative AMD but not PCV.
HTRA1 has its own role in both AMD and PCV24,25, governed by different molecular determinants. In this study, rs2672598, located in the promoter region of HTRA1, not only showed differential associations between AMD and PCV, but also altered the transcriptional activity of the HTRA1 promoter. Higher transcriptional activity of the HTRA1 promoter was observed in the constructs with rs2672598 C-allele rather than that with rs11200638 A-allele (Fig. 4), suggesting that the rs2672598 C-allele, not rs11200638 A-allele, is responsible for the alteration in the transcriptional activity of the HTRA1 promoter. Our luciferase-reporter analysis agreed with the transcription factor binding site prediction analysis (Fig. 5). No alteration of transcription factor binding site was found in rs11200638 A-allele when compared to the wildtype G-allele. In contrast, the transcription factor binding sites in rs2672598 C-allele were totally different from that in the wildtype T-allele. This further confirmed that rs2672598 should be the variant contributed to the altered transcriptional activity of the HTRA1 promoter. In order to validate the results of in vitro transcriptional activity, we performed a correlation analysis on the HTRA1 protein expression in vitreous humor with the rs2672598 genotypes. Our results showed that HTRA1 expression was higher in the vitreous humor with rs2672598 CC genotype than that with TT genotype by 2.56 folds (p = 0.02; Fig. 6). As we previously showed than rs11200638 is not correlated with HTRA1 expression in vitreous humor18, the vitreous humor expression results in this study not only validate our findings in the promoter analysis, but also confirm that rs2672598 is responsible for the higher HTRA1 expression in human ocular samples. Our luciferase activity and vitreous humor expression results of rs11200638 are similar to previous reports that there is no significant change of HTRA1 gene expression level in human retina among genotypes of HTRA1 rs11200638, rs1049331 and rs2293870, and ARMS2 rs10490924 and the del443ins54 variant26,27.
In summary, our results revealed that a HTRA1 promoter variant, rs2672598, is more significantly associated with exudative AMD than PCV. It should be responsible for elevated transcriptional activity in the HTRA1 promoter.
Methods
Study subjects
We recruited 188 PCV patients from the Prince of Wales Hospital Eye Centre and the Hong Kong Eye Hospital in Hong Kong. Together with 195 exudative AMD patients and 183 normal controls reported in our previous study28, a total of 534 unrelated Han Chinese were included (Supplementary Table 2). For validation, we recruited 187 PCV patients and 190 control subjects from the Joint Shantou International Eye Center of Shantou University and The Chinese University of Hong Kong, Shantou, China. All study subjects, including patients and controls, were given comprehensive ophthalmic evaluations and examined by clinical ophthalmologists. Both fluorescein angiography (FA) and ICGA were performed in the exudative AMD and PCV patients. AMD was graded according to an international classification and grading system29. Patients with exudative AMD had non-drusenoid RPE detachment, choroidal neovascularization, serous or hemorrhagic retinal detachments, sub-retinal or sub-RPE hemorrhage or fibrosis. PCV patients had sub-retinal red or orange nodules and hemorrhagic pigment epithelial detachment and characteristic sacculated vascular abnormalities in the inner choroid as visualized on ICGA. The diagnosis of PCV was distinguished from AMD by FA and ICGA staining30. PCV patients with geographic atrophy or early signs of AMD were excluded, together with patients who were difficult to be clearly differentiated (at late stage of disease that possess fibrosis and disciform scar or at advanced stage that present with intensive hemorrhage). The control subjects were recruited from elderly people greater than 60 years. They did not have any identifiable signs of AMD, PCV or other major eye diseases, except for mild senile cataract and slight refractive errors ranging from −1.5 to +1.5 diopters. The study protocol, approved by the Ethics Committee for Human Research at the Chinese University of Hong Kong, is in accordance with the tenets of the Declaration of Helsinki. Informed consent was obtained from each study subject.
Sequence analysis
Genomic DNA from whole blood was extracted (Qiagen QIAamp DNA Blood Mini kit, Qiagen, Hiden, Germany) according to the supplier’s instructions. The 9 exons, intron-exon junctions of HTRA1 (ENSG00000166033) and its promoter region were screened by polymerase chain reaction (PCR) with specific primers followed by direct sequencing (BigDye Terminator Cycle Sequencing Reaction Kit, v3.1; Applied Biosystems, Foster City, CA) on a DNA sequencer (ABI 3130XL, Applied Biosystems)28.
Association analysis
All the identified polymorphisms were assessed for Hardy-Weinberg equilibrium using χ2 analysis. Allelic and genotypic distributions for association between PCV and controls, between exudative AMD and controls, and between exudative AMD and PCV) were compared using the χ2 test or Fisher’s exact test (SPSS, version 16.0; SPSS Science, Chicago, IL). Logistic regression was used to examine the association profiles of the associated polymorphisms in HTRA1 between exudative AMD and PCV (SPSS). LD and haplotype-based association analyses were performed based on Haploview, version 4.2; http://www.broadinstitute.org/)31. Multiple testing correction was performed by permutation test (n = 10,000).
For the meta-analysis, the Review Manager software (RevMan, version 5.2, The Cochrane Collaboration, Copenhagen, Denmark) was used to generate the Z scores. The exact P values with the Z scores was then calculated using R (v3.0.0, http://cran.r-project.org/). The I2 statistic was used to assess the heterogeneity among studies, which corresponds to no (<25%), low (25–50%), moderate (50–75%), and high heterogeneity (≥75%). We adopted the random effects model in the meta-analysis. Summary p < 0.05 was considered statistically significant.
Joint genotype analysis
The genotype combinations of rs11200638 and rs2672598 in exudative AMD, PCV and control subjects were counted. Their corresponding odds ratios (OR) were calculated by the χ2 test (SPSS). The ORs were compared to the baseline genotype of the two genes that showed the lowest frequency of the disease risk alleles (homozygous G carrier of rs11200638 with homozygous T carrier of rs2672598).
Promoter analysis
A 726-bp genomic fragment (from −658 to +68) containing the HTRA1 promoter was cloned into empty pGL3-Basic vector, pGL3 (Promega, Madison, WI) between the NheI and BglII sites (OriGene Technologies, Rockville, MD). Genomic DNA of the study subjects were amplified by PCR (Ex Taq DNA Polymerase, TakaRa, Japan): Forward primer (5′-TAATGCTA GCTCTCTGCGAATACGGACACG), reverse primer (5′-TAATAGATCTGGGAGAGTGCAG GAGGG). Constructs with different rs11200638-rs2672598 haplotypes were generated. The cloned sequence of all constructs was verified by direct sequencing.
Human retinal pigment epithelial cell line ARPE-19 (American Type Culture Collection, Manassas, VA)32 was cultured in Dulbecco’s modified Eagle’s medium and F-12 nutrient mixture (DMEM/F-12) supplemented with 10% fetal bovine serum (Gibco BRL, Rockville, MD). Cells were plated in 60 mm tissue culture dishes at a density of 2–3 × 105 cells per dish one day before transfection. After 24 hours, cells were transfected with 2.5 μg of luciferase constructs in 7.5 μl FuGene HD transfection reagent (Roche, Indianapolis, IN) per dish. Empty pGL3 was used as negative control. At 72 hours after transfection, cell lysates were extracted by RIPA reagent (Sigma-Aldrich, St. Louis, MO) for detection of luciferase expression.
Luciferase expression was detected by immunoblotting according to our published method33. Briefly, the denatured cell lysates of the transfected cells were resolved on 10% SDS-polyacrylamide gel and electro-transferred to nitrocellulose membranes for probing with a mouse monoclonal primary antibody against firefly luciferase (LifeSpan BioSciences, Seattle, WA) and a secondary antibody against mouse IgG conjugated with horseradish peroxidase (Santa Cruz Biotechnology, Dallas, TX). Signals were detected by the enhanced chemiluminescence (ECL) system (Amersham Pharmacia, Cleveland, OH) and quantified by ChemiDoc (BioRad, Hercules, CA). Normalized luciferase intensities were calculated by dividing the quantified luciferase intensities by the housekeeping β-actin intensities. Two different clones of each haplotype were used in the transfection experiment. Triplicated experiments were performed. The wildtype rs11200638-rs2672598 haplotype (G-T) was set as a reference. One-way Analysis of Variance (ANOVA) with post-hoc tests adjusted by Tukey HSD (SPSS) was used to compare the means among different groups. Recombinant firefly luciferase (Promega) was used as a positive control.
DNA sequence of the cloned HTRA1 promoter was used to predict the transcription factor binding sites. We predicted the transcription factors that would bind to the region of rs11200638 and rs2672598 in the HTRA1 promoter by PROMO (version 3.0.2; http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3)34. Predictions with different alleles of rs11200638 and rs2672598 were performed.
Correlation analysis of rs2672598 genotype and HTRA1 expression in vitreous humor
Fifty-five unrelated Chinese patients underwent ocular surgeries at the Prince of Wales Hospital in Hong Kong and the Joint Shantou International Eye Center (JSIEC) of Shantou University and the Chinese University of Hong Kong were recruited and given complete ophthalmoscopic examinations18. In all patients, a standard three-port pars plana vitrectomy was performed as a part of the regular surgical procedures. Undiluted vitreous humor samples (0.5–1 ml) were collected into sterile tubes at the time of surgery and aliquots were rapidly frozen at −80 °C until assay.
HTRA1 protein expression has been previously determined18. Briefly, total protein concentrations in the vitreous samples were measured by Protein assay (BioRad, Hercules, CA). Equal amount of total protein (10 ug) for each denatured vitreous humor samples were resolved on 12.5% SDS-polyacrylamide gel and electro-transferred to nitrocellulose membranes for probing with the mouse monoclonal antibody against HTRA1 (R&D Systems Inc., Minneapolis, MN), and secondary antibody against mouse IgG conjugated with horseradish peroxidase (Jackson Immuno. Res., West Grove, PA). The signals were detected by enhanced chemiluminescence (ECL) (Amersham Pharmacia, Cleveland, OH) and the band intensities quantified by Quantity One® Image Analysis software (BioRad).
DNA was extracted from all of the vitreous humor samples using a commercially available DNA extraction kit (Qiagen, Germany). The HTRA1 rs2672598 genotypes were determined in all 55 samples by polymerase chain reaction (PCR) and direct DNA sequencing as described above28.
Additional Information
How to cite this article: Ng, T. K. et al. HTRA1 promoter variant differentiates polypoidal choroidal vasculopathy from exudative age-related macular degeneration. Sci. Rep. 6, 28639; doi: 10.1038/srep28639 (2016).
References
Pascolini, D. et al. 2002 global update of available data on visual impairment: a compilation of population-based prevalence studies. Ophthalmic Epidemiol 11, 67–115 (2004).
Jager, R. D., Mieler, W. F. & Miller, J. W. Age-related macular degeneration. N Engl J Med 358, 2606–2617 (2008).
Ciardella, A. P., Donsoff, I. M., Huang, S. J., Costa, D. L. & Yannuzzi, L. A. Polypoidal choroidal vasculopathy. Surv Ophthalmol 49, 25–37 (2004).
Ambati, J., Ambati, B. K., Yoo, S. H., Ianchulev, S. & Adamis, A. P. Age-related macular degeneration: etiology, pathogenesis, and therapeutic strategies. Surv Ophthalmol 48, 257–293 (2003).
Imamura, Y., Engelbert, M., Iida, T., Freund, K. B. & Yannuzzi, L. A. Polypoidal choroidal vasculopathy: a review. Surv Ophthalmol 55, 501–515 (2010).
Laude, A. et al. Polypoidal choroidal vasculopathy and neovascular age-related macular degeneration: same or different disease? Prog Retin Eye Res 29, 19–29 (2010).
Yannuzzi, L. A. et al. Polypoidal choroidal vasculopathy and neovascularized age-related macular degeneration. Arch Ophthalmol 117, 1503–1510 (1999).
Lai, T. Y., Chan, W. M., Liu, D. T., Luk, F. O. & Lam, D. S. Intravitreal bevacizumab (Avastin) with or without photodynamic therapy for the treatment of polypoidal choroidal vasculopathy. Br J Ophthalmol 92, 661–666 (2008).
Gomi, F. & Tano, Y. Polypoidal choroidal vasculopathy and treatments. Curr Opin Ophthalmol 19, 208–212 (2008).
Kwok, A. K., Lai, T. Y., Chan, C. W., Neoh, E. L. & Lam, D. S. Polypoidal choroidal vasculopathy in Chinese patients. Br J Ophthalmol 86, 892–897 (2002).
Honda, S. et al. Photodynamic therapy for typical age-related macular degeneration and polypoidal choroidal vasculopathy: a 30-month multicenter study in Hyogo, Japan. Jpn J Ophthalmol 53, 593–597 (2009).
Lim, J. Y. et al. Intravitreal bevacizumab alone versus in combination with photodynamic therapy for the treatment of neovascular maculopathy in patients aged 50 years or older: 1-year results of a prospective clinical study. Acta Ophthalmol 90, 61–67 (2012).
Maruko, I., Iida, T., Saito, M., Nagayama, D. & Saito, K. Clinical characteristics of exudative age-related macular degeneration in Japanese patients. Am J Ophthalmol 144, 15–22 (2007).
Nakata, I. et al. Significance of C2/CFB Variants in Age-related Macular Degeneration and Polypoidal Choroidal Vasculopathy in a Japanese Population. Invest Ophthalmol Vis Sci 53, 794–798 (2012).
Liang, X. Y. et al. Differentiation of exudative age-related macular degeneration and polypoidal choroidal vasculopathy in the ARMS2/HTRA1 locus. Invest Ophthalmol Vis Sci 53, 3175–3182 (2012).
Gotoh, N. et al. Haplotype analysis of the ARMS2/HTRA1 region in Japanese patients with typical neovascular age-related macular degeneration or polypoidal choroidal vasculopathy. Jpn J Ophthalmol 54, 609–614 (2010).
Cheng, Y. et al. Genetic and functional dissection of ARMS2 in age-related macular degeneration and polypoidal choroidal vasculopathy. PLoS One 8, e53665 (2013).
Ng, T. K. et al. Interactive expressions of HtrA1 and VEGF in human vitreous humors and fetal RPE cells. Invest Ophthalmol Vis Sci 52, 3706–3712 (2011).
Tong, J. P. et al. Aqueous humor levels of vascular endothelial growth factor and pigment epithelium-derived factor in polypoidal choroidal vasculopathy and choroidal neovascularization. Am J Ophthalmol 141, 456–462 (2006).
Kikuchi, M. et al. Elevated C-reactive protein levels in patients with polypoidal choroidal vasculopathy and patients with neovascular age-related macular degeneration. Ophthalmology 114, 1722–1727 (2007).
Matsumiya, W. et al. Early responses to intravitreal ranibizumab in typical neovascular age-related macular degeneration and polypoidal choroidal vasculopathy. J Ophthalmol 2011, 742020 (2011).
Kondo, N., Honda, S., Ishibashi, K., Tsukahara, Y. & Negi, A. LOC387715/HTRA1 variants in polypoidal choroidal vasculopathy and age-related macular degeneration in a Japanese population. Am J Ophthalmol 144, 608–612 (2007).
Lima, L. H. et al. Three major loci involved in age-related macular degeneration are also associated with polypoidal choroidal vasculopathy. Ophthalmology 117, 1567–1570 (2010).
Jones, A. et al. Increased expression of multifunctional serine protease, HTRA1, in retinal pigment epithelium induces polypoidal choroidal vasculopathy in mice. Proc Natl Acad Sci USA 108, 14578–14583 (2011).
Ng, T. K., Liang, X. Y. & Pang, C. P. HTRA1 in age-related macular degeneration. Asia Pac J Ophthalmol 1, 51–63 (2012).
Kanda, A. et al. Age-related macular degeneration-associated variants at chromosome 10q26 do not significantly alter ARMS2 and HTRA1 transcript levels in the human retina. Mol Vis 16, 1317–1323 (2010).
Wang, G. et al. Variants at chromosome 10q26 locus and the expression of HTRA1 in the retina. Exp Eye Res 112, 102–105 (2013).
Tam, P. O. et al. HTRA1 variants in exudative age-related macular degeneration and interactions with smoking and CFH. Invest Ophthalmol Vis Sci 49, 2357–2365 (2008).
Bird, A. C. et al. An international classification and grading system for age-related maculopathy and age-related macular degeneration, The International ARM Epidemiological Study Group. Surv Ophthalmol 39, 367–374 (1995).
Cackett, P., Wong, D. & Yeo, I. A classification system for polypoidal choroidal vasculopathy. Retina 29, 187–191 (2009).
Barrett, J. C., Fry, B., Maller, J. & Daly, M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005).
Dunn, K. C., Aotaki-Keen, A. E., Putkey, F. R. & Hjelmeland, L. M. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp Eye Res 62, 155–169 (1996).
Ng, T. K. et al. AC and AG dinucleotide repeats in the PAX6 P1 promoter are associated with high myopia. Mol Vis 15, 2239–2248 (2009).
Messeguer, X. et al. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18, 333–334 (2002).
Acknowledgements
We express our greatest appreciation to all the participants in the study. This study was supported by the Health and Medical Research Fund (project number: 12130791 to T.K.N.), the General Research Fund from the Research Grants Council (grant number: 473410 to C.P.P.), Hong Kong, and Direct Grants from the Medical Panel (grant number: 2041771 and 4054029 to C.P.P.), The Chinese University of Hong Kong.
Author information
Authors and Affiliations
Contributions
T.K.N., X.Y.L. and L.M. mainly performed the experiments. P.O.S.T. and J.X.W. helped with the experiments. T.Y.Y.L. and H.C. recruited study subjects. C.P.P. and L.J.C. supervised the project. T.K.N., X.Y.L. and C.P.P. interpreted the results and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Ng, T., Liang, X., Lai, T. et al. HTRA1 promoter variant differentiates polypoidal choroidal vasculopathy from exudative age-related macular degeneration. Sci Rep 6, 28639 (2016). https://doi.org/10.1038/srep28639
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep28639
This article is cited by
-
Contribution of common and rare variants to Asian neovascular age-related macular degeneration subtypes
Nature Communications (2023)
-
Hyaluronidase-1-mediated glycocalyx impairment underlies endothelial abnormalities in polypoidal choroidal vasculopathy
BMC Biology (2022)
-
Polymorphism rs11200638 enhanced HtrA1 responsiveness and expression are associated with age-related macular degeneration
Eye (2022)
-
Association of UCP1 and UCP2 variants with diabetic retinopathy susceptibility in type-2 diabetes mellitus patients: a meta-analysis
BMC Ophthalmology (2021)
-
HTRA1 rs11200638 variant and AMD risk from a comprehensive analysis about 15,316 subjects
BMC Medical Genetics (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.