Abstract
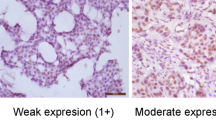
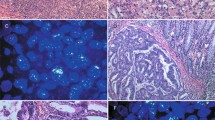
Tumor necrosis factor receptor (TNFR) associated factor 4 (TRAF4) was initially identified as a gene amplified and overexpressed in breast carcinomas. Our aim was to evaluate whether TRAF4 protein overexpression exists in other cancer types. Immunohistochemistry analysis of tumor samples from 623 patients with 20 different tumor types showed that TRAF4 was overexpressed in 268 tumors (43%), including 82 of 137 lung adenocarcinomas (60%). Interestingly, 32 primary tumors and their matching metastases exhibited mostly similar TRAF4 expression pattern. TRAF4 protein overexpression was limited to cancer cells and the subcellular localization was consistently cytoplasmic in a large majority of cases. To investigate changes in TRAF4 gene copy number, 125 cases from six different types of carcinomas were also analysed by fluorescence in situ hybridization. Out of the 28 cases (22%) showing an increased TRAF4 gene copy number, 23 (82%) were overexpressing the protein. Thus, TRAF4 gene amplification is one of the mechanisms responsible for TRAF4 protein overexpression in human cancers. Considering that TRAF4 is located at 17q11.2 in a region of amplification devoid of known oncogenes and is commonly overexpressed in cancer, our data support an oncogenic role for TRAF4.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Abell AN, Johnson GL . (2005). J Biol Chem 280: 35793–35796.
Aggarwal BB . (2003). Nat Rev Immunol 3: 745–756.
Balsara BR, Testa JR . (2002). Oncogene 21: 6877–6883.
Bieche I, Tomasetto C, Regnier CH, Moog-Lutz C, Rio MC, Lidereau R . (1996). Cancer Res 56: 3886–3890.
Bowers AJ, Boylan JF . (2004). Gene 328: 135–142.
Chung JY, Park YC, Ye H, Wu H . (2002). J Cell Sci 115: 679–688.
Collins C, Rommens JM, Kowbel D, Godfrey T, Tanner M, Hwang SI et al. (1998). Proc Natl Acad Sci USA 95: 8703–8708.
Esparza EM, Arch RH . (2004). Cell Mol Life Sci 61: 3087–3092.
Glauner H, Siegmund D, Motejadded H, Scheurich P, Henkler F, Janssen O et al. (2002). Eur J Biochem 269: 4819–4829.
Hirsch FR, Varella-Garcia M, Franklin WA, Veve R, Chen L, Helfrich B et al. (2002). Br J Cancer 86: 1449–1456.
Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E et al. (2002). Cancer Res 62: 6240–6245.
Kauraniemi P, Barlund M, Monni O, Kallioniemi A . (2001). Cancer Res 61: 8235–8240.
Krajewska M, Krajewski S, Zapata JM, Van Arsdale T, Gascoyne RD, Berern K et al. (1998). Am J Pathol 152: 1549–1561.
Li JM, Fan LM, Christie MR, Shah AM . (2005). Mol Cell Biol 25: 2320–2330.
Orsetti B, Nugoli M, Cervera N, Lasorsa L, Chuchana P, Ursule L et al. (2004). Cancer Res 64: 6453–6460.
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA et al. (2000). Nature 406: 747–752.
Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE et al. (2002). Proc Natl Acad Sci USA 99: 12963–12968.
Regnier CH, Masson R, Kedinger V, Textoris J, Stoll I, Chenard MP et al. (2002). Proc Natl Acad Sci USA 99: 5585–5590.
Regnier CH, Tomasetto C, Moog-Lutz C, Chenard MP, Wendling C, Basset P et al. (1995). J Biol Chem 270: 25715–25721.
Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D et al. (2004). Proc Natl Acad Sci USA 101: 9309–9314.
Sax JK, El-Deiry WS . (2003). J Biol Chem 278: 36435–36444.
Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE et al. (1989). Science 244: 707–712.
Takeshita F, Ishii KJ, Kobiyama K, Kojima Y, Coban C, Sasaki S et al. (2005). Eur J Immunol 35: 2477–2485.
Tomasetto C, Regnier C, Moog-Lutz C, Mattei MG, Chenard MP, Lidereau R et al. (1995). Genomics 28: 367–376.
Weigelt B, Glas AM, Wessels LF, Witteveen AT, Peterse JL, van't Veer LJ . (2003). Proc Natl Acad Sci USA 100: 15901–15905.
Wolf M, Mousses S, Hautaniemi S, Karhu R, Huusko P, Allinen M et al. (2004). Neoplasia 6: 240–247.
Wu RF, Xu YC, Ma Z, Nwariaku FE, Sarosi Jr GA, Terada LS . (2005). J Cell Biol 171: 893–904.
Xu YC, Wu RF, Gu Y, Yang YS, Yang MC, Nwariaku FE et al. (2002). J Biol Chem 277: 28051–28057.
Ye X, Mehlen P, Rabizadeh S, VanArsdale T, Zhang H, Shin H et al. (1999). J Biol Chem 274: 30202–30208.
Acknowledgements
We thank DJ Heard for critical reading of the manuscript, C Tomasetto for helpful discussions, F Devez and V Ducruit for expert technical assistance. The BAC clone RP11-386F9 was kindly provided by M Rocchi. This work was supported by funds from the Institut National de la Santé et de la Recherche Médicale, the Association pour la Recherche sur le Cancer (CH Régnier, contract no. 4801) and the Alliance des Recherches sur le Cancer.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Camilleri-Broët, S., Cremer, I., Marmey, B. et al. TRAF4 overexpression is a common characteristic of human carcinomas. Oncogene 26, 142–147 (2007). https://doi.org/10.1038/sj.onc.1209762
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1209762
Keywords
This article is cited by
-
TRAF6 as a potential target in advanced breast cancer: a systematic review, meta-analysis, and bioinformatics validation
Scientific Reports (2023)
-
SRC-3/TRAF4 facilitates ovarian cancer development by activating the PI3K/AKT signaling pathway
Medical Oncology (2023)
-
Stabilization of MCL-1 by E3 ligase TRAF4 confers radioresistance
Cell Death & Disease (2022)
-
Structural feature of TRAFs, their related human diseases and therapeutic intervention
Archives of Pharmacal Research (2021)
-
Ubiquitination of the DNA-damage checkpoint kinase CHK1 by TRAF4 is required for CHK1 activation
Journal of Hematology & Oncology (2020)