Abstract
Major depression is frequently associated with hyperactivity of the hypothalamic–pituitary–adrenal (HPA) axis. Clinically effective therapy with antidepressant drugs normalizes the disturbed activity of HPA axis, in part, by decreasing corticotropin-releasing hormone (CRH) synthesis, but the mechanism of this action is poorly recognized. In order to find out whether antidepressants directly affect CRH gene promoter activity, we studied their effect on undifferentiated and differentiated Neuro-2A cells, and for comparison the effect of the selected antidepressants on AtT-20 cells was also determined. The cells were stably transfected with a human CRH promoter fragment (−663 to +124 bp) linked to the chloramphenicol acetyltransferase (CAT) reporter gene. The regulation of CRH gene promoter activity is similar in Neuro-2A cells, both intact and differentiated, and in AtT-20 cell line, and cAMP/PKA-dependent pathway plays an important role in the stimulation of CRH gene. It was found that imipramine, amitryptyline, desipramine, fluoxetine, and mianserin, present in the culture medium for 5 days, in a concentration-dependent manner inhibited basal hCRH gene promoter activity in undifferentiated Neuro-2A cells, while other drugs under study (citalopram, tianeptine, moclobemide, venlafaxine, reboxetine, mirtazapine, and milnacipram) were inactive. In the differentiated cells, all examined antidepressants, except moclobemide (no effect) and tianeptine (increase), inhibited hCRH gene transcription. Moreover, in differentiated cells, the drugs acted stronger and were effective at lower concentrations. Forskolin-induced CAT activity was attenuated by imipramine and fluoxetine and to a lesser degree by amitriptyline and desipramine in differentiated cells, whereas other drugs were inactive. Moreover, imipramine and fluoxetine, but not tianeptine, showed moderate inhibitory effect on CRH gene promoter activity also in AtT-20 cell line, commonly used in CRH gene regulation studies. These results indicate that neuron-like differentiated Neuro-2A cells are a better model than pituitary and intact neuroblastoma to investigate the mechanism of psychotropic drug action. Inhibition of CRH gene promoter activity by antidepressant drugs may be a molecular mechanism by which these drugs inhibit the activity of HPA axis.
Similar content being viewed by others
INTRODUCTION
Dysregulation of the hypothalamic–pituitary–adrenocortical (HPA) axis is the most consistent biochemical change observed in patients suffering from major depression (Holsboer et al, 1985; Linkowski et al, 1985). High incidence of depression in Cushing's syndrome and antidepressant action of cortisol synthesis inhibitors and an antagonist of corticotropin-releasing hormone (CRH) receptors (Jeffcoate et al, 1979; Murphy 1997; O'Brien et al, 2001; Zobel et al, 2000) support the hypothesis that the hyperactivity of HPA is involved in the pathogenesis of this disorder. Moreover, the elevated CRH concentrations in the cerebrospinal fluid of depressed patients and increased number of CRH-expressing neurons in the hypothalamus (Nemeroff et al, 1984; Raadsheer et al, 1994) suggest that the hyperactivity of the HPA axis is a result of hypersecretion of CRH. In a transgenic mouse model, the increased production of CRH in the brain leads to disruption of dexamethasone-induced suppression of HPA activity. Interestingly, similar change has been observed in major depression (Groenink et al, 2002).
Antidepressant drugs are known to normalize HPA axis activity, at least partly, by decreasing CRH secretion. Some authors found that chronic administration of antidepressant drugs to animals decreased the CRH mRNA level in the rat hypothalamus; however, the lack of effect of some antidepressants on CRH synthesis under basal conditions was also reported (Brady et al, 1991, 1992; Stout et al, 2002). CRH-containing neurons primarily located in the parvocellular division of the paraventricular hypothalamic nucleus (PVN) are innervated by nerve fibers conveying impulses via numerous neurotransmitters and neuropeptides, which complicates elucidation of regulation of CRH synthesis and secretion under resting conditions and under various forms of stress. Most studies indicate that noradrenaline, serotonin, acetylcholine, histamine, and glutamate enhance the activity of HPA axis, whereas γ-aminobutyric acid and glucocorticoids act in the opposite direction (Herman and Cullinan, 1997). Antidepressant drugs are known to affect neurotransmitter and glucocorticoid receptor function, activities of various protein kinases, which may influence CRH gene expression. However, a molecular basis of interaction between antidepressants and CRH has not been recognized. This problem can be approached by investigation of the influence of antidepressant drugs on transcriptional regulation of the CRH gene in cultured cell lines. The sequence of the human CRH precursor gene has been determined (Shibahara et al, 1983) and regulation of its transcription has already been a subject of several investigations. The aim of our study was to find out if antidepressant drugs can directly affect human CRH gene transcription. The effects of those drugs were determined in cell cultures stably transfected with plasmid containing 663 bp of human CRH 5′-flanking DNA and 124 bp of the 5′-untranslated region of exon I linked to the chloramphenicol acetyltransferase (CAT) reporter gene. In the present study, we have chosen the Neuro-2A cell line, which displays a neuron-like phenotype both morphologically and neurochemically (Klebe and Ruddle, 1969). Since preliminary data showed that only few antidepressants affected CRH transcription in undifferentiated cells, we extended the study using differentiated Neuro-2A cells. Neuronal differentiation in vitro is accompanied by morphological alterations, including the development of neurites and restriction of cell division. The differentiated cells have higher intracellular level of serotonin, dopamine, and amino acids (Chatterjee et al, 1992; De Girolamo et al, 2000) and one might expect that they should be more sensitive to psychotropic drug action. Moreover, the regulation of CRH gene promoter activity and the effect of selected antidepressant drugs were also investigated in AtT-20 cell line, commonly used in CRH gene regulation studies. The transcription of CRH gene is positively regulated by cAMP/PKA-mediated mechanisms, and antidepressant drugs are known to affect this pathway (Guardiola-Diaz et al, 1994; Rossby et al, 1999; Schwaninger et al, 1995). Therefore, the effect of these drugs on forskolin-stimulated hCRH gene promoter activity was investigated. The following antidepressant drugs, with different neurochemical mechanisms of action, were investigated in the present study: imipramine and amitriptyline—nonselective serotonin (5-HT) and noradrenaline (NA) reuptake blockers; desipramine—a selective NA reuptake inhibitor, all exhibiting affinity for several neurotransmitter receptors; fluoxetine and citalopram—selective 5-HT reuptake blockers; tianeptine—a novel tricyclic drug, which increases the uptake of 5-HT; mianserin—an antagonist of α2-adrenergic receptors, which does not affect the uptake of the monoamines; moclobemide—a reversible monoamine oxidase-A inhibitor; venlafaxine and milnacipram—dual 5-HT and NA reuptake inhibitors, without effect on neurotransmitter receptors; reboxetine—the NA reuptake inhibitor without effect on neurotransmitter receptors; and mirtazapine—an antagonist of α2-adrenergic receptors. For the time-course study and for experiments with the AtT-20 cells, we selected three antidepressants, with quite different effects on monoamine uptake: imipramine—a nonselective 5-HT and NA uptake inhibitor and the most effective drug used in clinical practice; and fluoxetine and tianeptine, which show opposite effect on 5-HT uptake. Furthermore, since some antidepressant drugs used at high concentrations have been shown to induce cell death in some neuronal cell lines (Post et al, 2000), their effects on Neuro-2A cell viability were studied.
Preliminary data on the effects of imipramine, fluoxetine, and tianeptine on CRH gene promoter activity in intact Neuro-2A cells have been published (Budziszewska et al, 2002).
METHODS
Cell Culture Conditions
Neuro-2A (mouse neuroblastoma) and AtT-20/D16v-F2 (mouse pituitary) cells were obtained from the American Type Culture Corporation (ATCC). Neuro-2A were cultured in Dulbecco's modified Eagle's medium (Gibco BRL) supplemented with 10% fetal bovine serum (Gibco BRL), 50 U/ml of penicillin, and 50 μg/ml of streptomycin (Sigma Co.), in an atmosphere composed of 5% CO2/95% O2 at 37°C. For in vitro differentiation, the cells were grown for 3 days before experiment in medium containing 1% serum. AtT-20/D16v-F2 cells were maintained in nutrient mixture Ham's F-10 medium (Gibco BRL) supplemented with 25 mM glucose, 15% horse serum (Gibco BRL), 2.5% fetal bovine serum, 50 U/ml of penicillin, and 50 μg/ml of streptomycin (Sigma Co.).
Plasmid Construction
hCRHCAT plasmid was synthesized as described previously (Budziszewska et al, 2002). Human DNA was isolated from blood using the Blood DNA Prep Kit (A&A Biotechnology, Poland). Based on the known human CRH gene promoter sequence (GenBank accession no. AF48855), the specific primers from the region −663 to +124 bp were synthesized for PCR amplification. The forward primer was CRHKpn 5′ CGC GGT ACC GAG AGA CGT CTC CGG GGG C 3′ (28 nt, containing a KpnI recognition site), and reverse primer was CRHBgl 5′ GCG AGA TCT GGC TCA TAA CTC CTT TAT GTG CTT GC 3′) (35 nt, containing a BglII recognition site). The bold parts of primer sequences are complementary to the nucleotide sequences of CRH promoter, whereas 5′ overhanging ends of primers contain recognition sites for restriction endonucleases (underlined), and serve to facilitate cloning. The reaction mixture consisted of: 50 ng of the human genomic DNA, 2 μl (10 μM) of each primer, 5 μl (10 mM) of dNTPs, 5 μl of 10 × PCR buffer (100 mM Tris-HCl, pH 8.9, 500 mM KCl, 20 mM MgCl2, 1% Triton X-100), and 2 U of Pwo DNA polymerase (Dąbrowski and Kur, 1998). The fragment was amplified by seven cycles performed with the following temperature profile: 30 s at 94°C, 1 min at 58°C, and 1 min at 72°C, and then 33 cycles (30 s at 94°C, 1 min at 70°C, and 1 min at 72°C). The amplification product was analyzed by electrophoresis on a 1% agarose gel stained with ethidium bromide. Specific, approximately 800 bp long, PCR product was obtained (0.2 μg), which was purified using the Clean-Up Kit (A&A Biotechnology, Poland), and digested with BglII and KpnI, and then isolated from an agarose gel band using the Gel-Out Kit (A&A Biotechnology, Poland). The purified fragment was ligated into pSecTag2A (Invitrogen, USA) BglII–KpnI sites. Escherichia coli TOP10F′ cells (Invitrogen, USA) were transformed with the ligation mixture, and 12 colonies were assayed for the presence of CRH promoter encoding gene by PCR amplification and restriction analysis. Four independent clones with CRH promoter were selected and sequenced, and compared to the known DNA sequence. After sequencing, one plasmid was selected, named pCRH, and used for next construct. Gene encoding CAT was amplified using primers CATBamHI 5′ GCG GGA TCC ATG GAG AAA AAA ATC ACT GG 3′ (29 nt, containing a BamHI recognition site) and CATXhoI 5′ CGC CTC GAG TTA CGC CCC GCC CTG CC 3′ (26 nt, containing a XhoI recognition site). pACYC184 plasmid (2 ng) was used as a source of DNA containing CAT gene. The product was amplified by seven cycles (30 s at 94°C, 1 min at 50°C, and 1 min at 72°C), and then 33 cycles with the following temperature profile: 30 s at 94°C, 1 min at 65°C, and 1 min at 72°C. Specific PCR product (666 bp) was obtained, purified, and digested with BamHI and XhoI, and ligated into pCRH BamHI–XhoI plasmid sites. The obtained pCRHCAT plasmid was analyzed by restriction digestion and sequencing. Schematic construction of an hCRHCAT fusion gene is shown in Figure 1.
Schematic representation of hCRHCAT fusion gene. PCRH: human CRH promoter gene (−663/+124 bp); CAT: coding region of the CAT gene; BGHpA: bovine growth hormone polyadenylation and termination signals; F1ori: F1 origin; SV40 ⇒ : SV40 early promoter and origin; ZeoR: zeocin resistance gene; SV40: polyadenylation signal; ColE1: fragment allowing high-copy number replication and growth in E. coli; AMPR: ampicillin resistance gene.
Cell Transfection
Cells at 50–60% confluence were transfected with hCRHCAT plasmid using LipofectAMINE reagent (Invitrogen, USA) according to recommendations of the manufacturer. Briefly, the cells were plated 1 day prior to transfection in a six-well tissue culture dish and grown in a medium with serum, but without antibiotics. Just before transfection, the cells were washed once with 2 ml of serum-free medium (Opti- MEM I; Invitrogen, USA). For each well, 12 μl of LipofectAMINE Reagent was diluted in 100 μl Opti-MEM I and incubated at room temperature for 30 min. Next, 1 μg of DNA diluted in 100 μl Opti-MEM I was added to LipofectAMINE solution, and after 30 min incubation at room temperature, DNA–liposome complexes, diluted with 0.8 ml of Opti-MEM I, were added to the cells. Cells were incubated for 6 h in the incubator, and then 1 ml of growth medium containing twice the normal concentration of serum was added. At 24 h after the beginning of transfection, the medium with lipid–DNA complex was replaced by a fresh, normal growth medium. Because pSecTag2A vector contains zeocin-resistant gene, at 72 h after transfection, the cells were transferred (1 : 6) to the selective medium (with 600 μg/ml of zeocin for Neuro-2A cell line and 200 μg/ml of zeocin for AtT-20 cells; Invitrogen, USA). Before transfection, the dose–response assays were performed, which showed that 300 and 100 μg/ml of zeocin were the lowest concentrations that killed all nontransfected Neuro-2A and AtT-20 cells, respectively. At 3–4 weeks after transfection, the zeocin-resistant colonies were selected, cultured in the medium with zeocin (150 μg/ml for Neuro-2A and 50 μg/ml for AtT-20 cells) and assayed for reporter gene activity. Neuro-2A cell line transfected with the promoter-less vector (pCAT3-basic, Invitrogen, USA) was used as negative control. The plasmid that contained human cytomegalovirus (hCMV) instead of CRH promoter was used as the control for antidepressant drug action.
Drug Treatments
In the first experiment, the regulation of CRH gene promoter activity in both cell lines was investigated. Stably transfected Neuro-2A cells (intact and differentiated) and stably transfected AtT-20/D16-F2 cells (final confluence of 70%) were treated with the appropriate vehicle, 25 μM forskolin (Calbiochem), 0.1 μM phorbol 12-myristate 13-acetate (TPA, RBI), 50 mM aqueous solution of KCl, and 1 μM dexamethasone (Sigma, St Louis, USA) for time periods indicated in the Results section. Forskolin and TPA were dissolved in DMSO (final concentration of DMSO was 0.5%), while dexamethasone was dissolved in a small amount of ethanol, followed by dilution in water (final concentration of ethanol was below 0.5%). The control cultures were supplemented with the same amount of an appropriate vehicle.
In the next experiment, the influence of antidepressant drugs on basal CRH gene promoter activity in Neuro-2A cells was determined. The cells were cultured in the presence of vehicle, imipramine hydrochloride (Polfa, Poland), amitriptyline hydrochloride (Polfa, Poland), desipramine hydrochloride (RBI, Natick, USA), fluoxetine hydrochloride (Eli Lilly, USA), citalopram hydrobromide (Lundbeck, Denmark), tianeptine hydrochloride (Servier, France), mianserin hydrochloride (Organon, The Netherlands), moclobemide (Hoffman-La-Roche, Switzerland), venlafaxine hydrochloride (Wyeth-Ayerst, Princeton, USA), reboxetine hydrochloride (Pharmacia–Upjohn, USA), mirtazapine (Organon, The Netherlands), and milnacipram hydrochloride at concentrations of 3, 10, 30 μM (intact cells) and 1, 3, 5 μM (differentiated cells) for 5 days. In the next phase of the experiment, the influence of all the above-listed antidepressant drugs at the same concentrations present in the medium for 5 days on forskolin-stimulated CAT activity was measured in intact and differentiated Neuro-2A cells. Forskolin (25 μM) was added to culture medium 24 h before harvesting the cells. Antidepressant drugs were dissolved in water and control cultures were supplemented with the same amount of water. The medium and drugs were changed once during 5-day culture.
In the time-course study, the effect of imipramine, fluoxetine, and tianeptine (3, 10, and 30 μM) on basal CRH gene promoter activity in intact Neuro-2A cells was determined after culturing the cells in the drug-supplemented medium for 5, 24, 48, and 72 h. Furthermore, the effect of the selected antidepressant drugs (imipramine, fluoxetine, and tianeptine) at 3, 10, and 30 μM concentrations on basal and forskolin (25 μM, 24 h)-induced CAT activity was studied in AtT-20 cells cultured in the presence of the drugs for 5 days.
CAT Activity
CAT activity was determined as described previously (Budziszewska et al, 2000; Pariante et al, 1997). Cell lysates were prepared by a freezing/thawing procedure. To determine CAT activity, aliquots of lysate (after heating at 60°C for 10 min) were incubated in 0.25 M Tris-HCl buffer (pH 7.8) with 0.25 μCi D-threo-[dichloroacetyl-1-14C]-chloramphenicol and 0.2 mM n-butyryl coenzyme A at 37°C for 1 h. The butyrylated forms of chloramphenicol (in direct proportion to the CAT gene expression) were extracted twice with xylene, washed with 0.25 M Tris-HCl buffer, and radioactivity was measured in a β-counter (Beckmann LS 335 liquid scintillation counter). The results are presented as dpm of a butyrylated fraction of chloramphenicol per 100 μg of protein per 1 h of incubation. The protein concentration in cell lysates was determined by the method of Lowry.
Effect of Antidepressant Drugs on Cell Viability
Neuro-2A cells were treated with the vehicle or antidepressant drugs at final concentrations of 3, 10, and 30 μM for 5 days. The effect of drugs on cell viability was determined by counting viable and nonviable (blue) cells in a hemocytometer. The cell suspensions were mixed (at a 1 : 1 ratio) with 0.4% trypan blue, and the number of nonviable cells per a total of 100 cells was calculated.
Statistical Analysis
The data are presented as means±SEM of three to five independent experiments (in duplicate wells), and the significance of differences between the means was evaluated by the Duncan's test following one-way or two-way analysis of variance, respectively.
RESULTS
The Regulation of CRH Gene Promoter Activity
In order to study the regulation of CRH gene promoter activity, the Neuro-2A and AtT-20/D16v cells were stably transfected with a fusion human CRH reporter gene. Several independent zeocin-resistant clones were isolated and tested for basal and forskolin-induced CAT activity, to find possible differences resulting from integration of the plasmid at different sites in the cell genome. Treatment of cells with 25 μM forskolin, an activator of adenylate cyclase, increased the CAT activity (2- to 6-fold) in 38/40 of the examined Neuro-2A subclones and in 17/20 of AtT-20. For further studies, we chose subclones with typical basal and forskolin-induced CAT activity.
Treatment of stably transfected intact Neuro-2A cells with 25 μM forskolin increased the CAT activity in a time-dependent manner (Figure 2). TPA, an activator of protein kinase C, had no effect on reporter gene activity, but enhanced the forskolin effect when it was present in the medium for 24 h. Similarly, KCl given alone at the concentration that induces membrane depolarization (50 mM) had no effect on CRH gene promoter activity, but strongly potentiated the forskolin action. Dexamethasone (1 μM) present in culture medium for 24 h had no effect on basal CAT activity, but attenuated the effect of forskolin. In differentiated Neuro-2A cells, forskolin (25 μM, 24 h) increased CAT activity 2–3 times (Table 2), while TPA (0.1 μM, 24 h) and dexamethasone (1 μM, 24 h) alone had no effect, but dexamethasone inhibited forskolin action by about 45% (data not shown). Similar regulation of CRH gene promoter activity as in Neuro-2A cells was found in AtT-20/D16v-F2 cells. Forskolin (25 μM, 24 h) increased CAT activity about three-fold (Figure 5), whereas dexamethasone (1 μM, 24 h) did not change basal CAT activity, but attenuated (by 30%) forskolin-induced reporter gene. TPA (0.1 μM, 24 h) and KCl (50 mM, 24 h) had no effect on basal and forskolin-induced CAT activity; however, a tendency to increase forskolin action was observed (data not shown).
Effect of forskolin, KCl, phorbol 12-myristate 13-acetate (TPA), and dexamethasone (Dexa) applied at the indicated concentrations and time on the CAT gene transcription in Neuro-2A cells stably transfected with pCRHCAT plasmid. The data are calculated as the dpm of the butyrylated fraction of chloramphenicol per 100 μg of protein per 1 h of incubation and presented as a percentage±SEM of control (culture with appropriate vehicle only). The significance of differences between the means was evaluated by the Dunnett's test following a one-way analysis of variance (*p<0.01 vs control group; # p<0.01 vs forskolin-treated group).
Effect of imipramine, fluoxetine, and tianeptine applied at the indicated concentrations for 5 days on the basal and forskolin (25 μM, 24 h)-induced CAT gene transcription in AtT-20 cells stably transfected with pCRHCAT plasmid. The data are calculated as the dpm of the butyrylated fraction of chloramphenicol per 100 μg of protein per 1 h of incubation and presented as a percentage±SEM of control (culture with appropriate vehicle only). The significance of differences between the means was evaluated by the Dunnett's test following a one-way analysis of variance (*p<0.01 vs control group).
Effect of Antidepressant Drugs on Basal CRH Gene Promoter Activity in Intact Neuro-2A Cells
Treatment of the cells with imipramine, fluoxetine, and tianeptine at concentrations of 3, 10, and 30 μM for 5, 24, 48, and 72 h had no statistically significant effect on basal CAT activity; however, a decreasing tendency was observed after 72 h (data not shown). The first significant effects appeared after 5 days of the presence of some antidepressant drugs in culture medium. Imipramine, amitriptyline, desipramine, fluoxetine, and mianserin in a concentration-dependent manner inhibited CAT activity, while other drugs under study, that is, citalopram, tianeptine, moclobemide, venlafaxine, reboxetine, mirtazapine, and milnacipram, had no effect (Table 1). No drug used at the above concentrations had any effect on CAT activity in Neuro-2A cells transfected with the promoter-less vector or transfected with plasmid containing CMV instead of CRH promoter (data not shown).
Effect of Antidepressant Drugs on Basal CRH Gene Promoter Activity in Differentiated Neuro-2A Cells
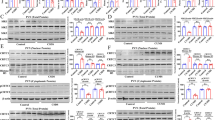
Neuro-2A cells grown in the medium containing 1% serum proliferated slightly, and about 80% of the cells had differentiated, for example formed long neurites. Imipramine, amitriptyline, desipramine, fluoxetine, citalopram, mianserin, venlafaxine, reboxetine, mirtazapine, and milnacipram in a concentration-dependent manner inhibited CRH gene promoter activity in differentiated Neuro-2A cells (Figure 3 and 4). Moclobemide had no effect, while tianeptine at the highest concentration used (5 μM) increased CAT activity.
Effect of antidepressant drugs applied at the indicated concentrations for 5 days on the CAT gene transcription in differentiated Neuro-2A cells stably transfected with pCRHCAT plasmid. The data are calculated as the dpm of the butyrylated fraction of chloramphenicol per 100 μg of protein per 1 h of incubation and presented as a percentage±SEM of control (culture with appropriate vehicle only). The significance of differences between the means was evaluated by the Dunnett's test following a one-way analysis of variance (*p<0.01 vs control group).
Effect of antidepressant drugs applied at the indicated concentrations for 5 days on the CAT gene transcription in differentiated Neuro-2A cells stably transfected with pCRHCAT plasmid. The data are calculated as the dpm of the butyrylated fraction of chloramphenicol per 100 μg of protein per 1 h of incubation and presented as a percentage±SEM of control (culture with appropriate vehicle only). The significance of differences between the means was evaluated by the Dunnett's test following a one-way analysis of variance (*p<0.01 vs control group).
Effect of Antidepressant Drugs on Forskolin-Induced CRH Gene Promoter Activity in Intact and Differentiated Neuro-2A Cells
Forskolin (25 μM), present in culture medium for 24 h, increased about three-fold the CAT activity in intact and differentiated Neuro-2A cells. Treatment of intact cells with imipramine (10 and 30 μM), amitriptyline (30 μM), and desipramine (30 μM) decreased forskolin-induced CAT activity (Table 2), while other drugs under study, that is, fluoxetine, citalopram, tianeptine, mianserin, moclobemide, venlafaxine, reboxetine, mirtazapine, and milnacipram, had no effect (data not shown). In differentiated cells, imipramine and fluoxetine (3 and 5 μM) strongly inhibited CAT activity, amitriptyline and desipramine (5 μM) had less potent effect (Table 2), while other drugs under study were without significant effect (data not shown).
Effect of Imipramine, Fluoxetine, and Tianeptine on Basal and Forskolin-Induced CRH Gene Promoter Activity in AtT-20/D16v-F2 Cells
Imipramine (10 and 30 μM) and fluoxetine (30 μM), but not tianeptine, present in the medium for 5 days inhibited the basal CAT activity, whereas forskolin action was attenuated only by 30 μM imipramine (Figure 5).
Effect of Antidepressant Drugs on Cell Viability
None of antidepressant drugs used in the present study (except desipramine and fluoxetine used at 30 μM concentration) evoked toxic effect, estimated by counting nonviable cells, in intact Neuro-2A cells. Desipramine and fluoxetine used at the highest concentration increased the amount of nonviable cells by 30 and 20%, respectively.
DISCUSSION
In the first part of the present study, the basic regulatory mechanism of CRH gene expression was investigated. The influence of PKA and PKC activation, membrane depolarization and dexamethasone on CRH promoter activity in Neuro-2A cells, well-characterized cells that display a neuron-like phenotype both morphologically and neurochemically (Chatterjee et al, 1992; Kadota et al, 1997; Klebe and Ruddle 1969), was determined and compared with the results obtained in AtT-20 (pituitary) cell line, commonly used to study CRH gene regulation. It has been found that CRH gene transcription is positively regulated by forskolin (a cAMP/PKA pathway activator) in both cell types. This finding supports and extends previous reports showing that cAMP-dependent regulation of hCRH promoter can be observed in AtT-20, PC-12, and human neuroblastoma (SK-N-MC) cell cultures, but not in cells with low levels of endogenous cyclic AMP-response element binding protein (CBP) (Guardiola-Diaz et al, 1994, 1996; Rosen et al, 1992; Seasholtz et al, 1988; Spengler et al, 1992; Van et al, 1990). These data are in agreement with in vivo study, which demonstrated that microinjection of 8-Br-cAMP into the hypothalamus increased CRH synthesis (Itoi et al, 1996), and with the report confirming the presence of the functional CRE sequence upstream of the regulatory region of the CRH gene (Seasholtz et al, 1988). In contrast to forskolin, the activator of protein kinase C (TPA) had no effect on CRH gene promoter activity in both cell lines studied, which suggests that the phospholipase C/protein kinase C (PLC/PKC) pathway is not involved in the regulation of human CRH gene. In fact, the absence of a clearly discernible perfect TPA- responsive element in the proximal 0.9-kb 5′ flanking the hCRH gene has been reported (Vamvakopoulos and Chrousos, 1994), and microinjection of TPA into the hypothalamus did not change CRH synthesis (Itoi et al, 1996). Like TPA, also membrane depolarization evoked by potassium did not change basal CRH promoter activity, but enhanced forskolin action in Neuro-2A cells and showed similar tendency in AtT-20 cells. Although molecular interactions between transcription factors are poorly understood yet, enhancement of forskolin action by TPA may result from heterodimerization of CREB/ATF and AP-1 family members at the CRE element (Malkoski and Dorin, 1999). Synergistic activation of CRH promoter activity by potassium and forskolin, previously found also in PC-12 cells (Guardiola-Diaz et al, 1994), is an example of frequently observed convergent effects of depolarization and cAMP on CRE-mediated gene expression. It has been postulated that glucocorticoids inhibit CRH gene promoter activity via interference with the cAMP-mediated activation (Guardiola-Diaz et al, 1996). Indeed, dexamethasone caused a moderate suppression of forskolin-induced CRH promoter activity in both Neuro-2A and AtT-20 cultures, which is in line with the previous study conducted on the latter cells (Van et al, 1990). However, lack of dexamethasone effect on basal CRH gene promoter activity and weaker action of corticosterone than dexamethasone on forskolin-induced action (Woods et al, 1992; our unpublished observation) were unexpected. Relatively weak action of glucocorticoids on CRH gene expression in cell lines could be due to the absence of mineralocorticoid receptors (MR), which have been shown in in vivo studies to inhibit basal and, together with GR, the stress-induced HPA axis activity (Ratka et al, 1989). The weaker action of corticosterone than dexamethasone might result also from fast metabolism of the former (Woods et al, 1992; our unpublished observation).
In the second part of this study, the effect of various antidepressant drugs on CRH gene promoter activity in Neuro-2A cell line was determined. The main finding of these experiments is the demonstration that 10 from among 12 antidepressant drugs under study are able to inhibit basal hCRH gene promoter activity in differentiated Neuro-2A cells. Also, in intact Neuro-2A cells, some antidepressant drugs, that is, imipramine, amitryptyline, desipramine, fluoxetine, and mianserin, inhibit basal hCRH gene promoter activity. However, in differentiated cells, the drugs acted stronger and were effective at lower concentrations. Inhibitory effect of imipramine and fluoxetine was also observed in AtT-20 cells. Consistent with these data are reports that long-term administration of some antidepressant drugs decreases CRH mRNA levels in the rat hypothalamus (Aubry et al, 1999; Brady et al, 1991, 1992; Fadda et al, 1995). The data on fluoxetine effect on CRH synthesis and HPA axis activity are not unequivocal (Brady et al, 1992; Stout et al, 2002); however, our results indicate that fluoxetine is able to inhibit CRH gene expression at least under in vitro conditions. Assuming similar regulation of CRH gene transcription in Neuro-2A cells and hypothalamic neurons, the inhibitory effect of antidepressants on CRH gene promoter activity may be a molecular mechanism by which these drugs inhibit CRH synthesis. In contrast to a majority of antidepressants under study, moclobemide had no effect on CRH gene promoter activity in both intact and differentiated Neuro-2A cells. This is in line with other reports that showed that moclobemide did not affect gene transcription in various experimental models (Post et al, 2000; Schwaninger et al, 1995). Although it is not known whether the effect of antidepressants on CRH gene transcription is connected with their effect on monoamine levels, it is possible that lack of moclobemide effect resulted from low MAO activity in neuroblastoma cells (Skaper et al, 1976). On the other hand, tianeptine, a new atypical antidepressant drug, which acts oppositely to other antidepressants on 5-HT uptake, enhanced CRH gene promoter activity in differentiated Neuro-2A cells. Stimulatory effect of tianeptine is unexpected, since among antidepressants tianeptine most effectively blocked various consequences of chronic stress. However, its effect on HPA axis activity varies depending on experimental paradigm. It has been reported that tianeptine decreased, had no effect, or even increased HPA axis activity (Delbende et al, 1994; Kole et al, 2002; Watanabe et al, 1992). Recent data indicate that effect of tianeptine on stress-evoked changes is not connected with its action on corticosterone level (Kole et al, 2002), and that this drug can directly inhibit glucocorticoid receptor-mediated gene transcription (Budziszewska et al, 2000). Thus, it is not unlikely that at least the stimulatory effect of tianeptine on HPA axis activity (Kole et al, 2002) may involve its direct effect on CRH gene promoter activity, especially when this drug is used at high concentration.
At present it is difficult to predict what kind of mechanism could explain the inhibitory action of most of the antidepressants on basal CRH gene promoter activity. Antidepressant drugs differ among themselves with respect to their influence on monoamine transmission. Neuro-2A cells contain 5-HT and 5-HT transporter, whose level increased during differentiation; however, they possess very little NA (Chatterjee et al, 1992). Opposite regulation of CRH gene transcription by tianeptine and fluoxetine, the drugs that enhanced and decreased intracellular 5-TH concentration, respectively, may suggest a functional role of this monoamine in CRH regulation. On the other hand, both selective inhibitors of NA uptake (desipramine and reboxetine) as well as drugs that do not affect monoamine uptake (mianserin and mirtazapine) showed also high activity in the present study. Thus, although the role of 5-HT cannot be excluded, it seems rather unlikely that monoamines play an important role in all antidepressants effects on CRH gene transcription.
Antidepressants are believed to inhibit HPA axis activity in vivo by increasing glucocorticoid receptors (GR) level in the rat central nervous system, which leads to enhancement of the GR-mediated feedback inhibition (Pariante and Miller, 2001). Some antidepressant drugs could also directly inhibit GR-mediated gene transcription under in vitro condition (Budziszewska et al, 2000; Okuyama-Tamura et al, 2003). However, action on GR level or GR-induced inhibition of CRH transcription does not seem to be the mechanism responsible for the action of antidepressants in the present study. As we showed, dexamethasone or corticosterone did not inhibit basal CRH gene promoter activity. Furthermore, inhibitory effect of dexamethasone on forskolin-induced CRH gene transcription was not enhanced by antidepressants (our unpublished data).
Alternatively, the direct effect of antidepressants on the activity of some protein kinases may be responsible for inhibition of CRH gene transcription. Antidepressant drugs are known to modulate the activity of various protein kinases by direct and receptor-mediated mechanism (Nibuya et al, 1996; Rossby et al, 1999; Schwaninger et al, 1995). The cAMP/PKA pathway potently stimulates CRH promoter activity, but the involvement of PKA as well as other protein kinases in the regulation of basal CRH gene transcription was not studied yet. Our preliminary data showed that an inhibitor of PKA lowered basal CRH gene promoter activity. Antidepressant drugs are believed to enhance PKA activity; however, the fact that both antidepressant drugs and PKA inhibitor reduced basal CRH activity suggested that if PKA was involved in the action of antidepressants on CRH, these drugs inhibited its activity. Moreover, some antidepressants reduced forskolin action on CRH promoter activity confirming their inhibitory effect on PKA activity. However, involvement of PKA and other protein kinases in the action of antidepressant drugs on CRH transcription will be a subject of further research.
Since the cAMP/PKA signal transduction is the main pathway that stimulates CRH gene promoter activity, the effect of antidepressant drugs on forskolin-induced CAT activity was also evaluated in the present study. Among drugs under study, only imipramine, fluoxetine, and higher concentrations of amitriptyline and desipramine were able to inhibit forskolin-induced CRH gene promoter activity in differentiated Neuro-2A cells. Inhibition of forskolin-induced CRH gene transcription by some antidepressant drugs is in line with the data indicating that expression of some genes containing CRE in their promoter region is downregulated in cell cultures (Schwaninger et al, 1995) and in the rat brain after chronic treatment with antidepressants (Brady et al, 1991; Hosoda and Duman 1993; Nestler et al, 1990). Although prevailing evidence indicates that antidepressants enhance cAMP/PKA pathway and increase the level of CREB in the rat brain (Nibuya et al, 1996), some recent reports indicate that they may inhibit the level of transcription factor, phospho-CREB. In fact, it has been shown that phospho-CREB is downregulated in the rat frontal cortex by chronic administration of desipramine, reboxetine, imipramine, and venlafaxine (Manier et al, 2002; Rossby et al, 1999; Tamura et al, 2002). A question arises, why reboxetine and venlafaxine, which decreased phospho-CREB in the above-cited experiments, were inactive in our study? It can only be speculated that their intracellular concentration after 5-day culture is sufficient to affect basal CRH gene transcription, but it may be too low to influence the activity of phospho-CREB. We used antidepressant drugs at concentrations that should mimic their brain therapeutic level. The therapeutic concentrations of tricyclic antidepressants range between 0.3 and 1 μM in the plasma, and are about 20–30 times higher in the rat brain (Glotzbach and Preskorn, 1982; Hrdina and Dubas, 1981; Miyake et al, 1990). Although precise concentration of antidepressants in the human brain is largely unknown, and the estimates are based on animal studies and few reports on detections of psychoactive drugs (with a trifluoromethyl group) in the human head by nuclear magnetic resonance spectroscopy (Bartels and Albert, 1995; Karson et al, 1992; Komoroski et al, 1994), the concentrations between 3 and 30 μM should approximate their therapeutic brain concentrations. For example, fluoxetine concentrations in the human brain are between 3.8 and 29.5 μM (Karson et al, 1992; Komoroski et al, 1994). Imipramine, amitryptyline, desipramine, and fluoxetine, the drugs active in the experiments with forskolin and in the study on basal CRH activity in undifferentiated Neuro-2A cells, are weak bases (pKa: from 9.4 to 10.5), and are known to penetrate and accumulate in the cells (Altamura et al, 1987; Kornhuber et al, 1995). However, lack of any data on intracellular concentration of some new antidepressant drugs, especially in the nuclear fraction, does not permit any general conclusion in this matter. In the light of the critical role of the CRE sequence in the regulation of CRH gene (Guardiola-Diaz et al, 1994), our data suggest that stress-induced increase in CRH gene transcription should be inhibited mainly by tricyclic antidepressant drugs.
In conclusion, the present data indicate that the regulation of CRH gene promoter activity is similar in Neuro-2A and AtT-20 cell lines and that cAMP/PKA-dependent pathway plays an important role in the stimulation of CRH gene. Stronger inhibitory action of antidepressants on CRH gene in differentiated Neuro-2A cells suggests that neuron-like cells may be a better model than pituitary or intact neuroblastoma cells to investigate the mechanism of action of psychotropic drugs. The ability of antidepressant drugs to inhibit basal and, to a lesser extent, the forskolin-induced activity of CRH gene transcription may be a molecular mechanism involved in their inhibitory effect on HPA axis activity.
References
Altamura AC, De Novellis F, Mauri MC, Gomeni R (1987). Plasma and brain pharmacokinetics of mianserin after single and multiple dosing in mice. Prog Neuropsychopharmacol Biol Psychiatry 11: 23–33.
Aubry JM, Pozzoli G, Vale WW (1999). Chronic treatment with the antidepressant amitriptyline decreases CRF-R1 receptor mRNA levels in the rat amygdala. Neurosci Lett 266: 197–200.
Bartels M, Albert K (1995). Detection of psychoactive drugs using 19F MR spectroscopy. J Neural Transm Gen Sect 99: 1–6.
Brady LS, Gold PW, Herkenham M, Lynn AB, Whitfield HJ (1992). The antidepressants fluoxetine, idazoxan and phenelzine alter corticotropin-releasing hormone and tyrosine hydroxylase mRNA levels in rat brain: therapeutic implications. Brain Res 572: 117–125.
Brady LS, Whitfield HJ, Fox RJ, Gold PW, Herkenham M (1991). Long-term antidepressant administration alters corticotropin-releasing hormone, tyrosine hydroxylase, and mineralocorticoid receptor gene expression in rat brain. J Clin Invest 87: 831–837.
Budziszewska B, Jaworska-Feil L, Kajta M, Lasoñ W (2000). Antidepressant drugs inhibit glucocorticoid receptor-mediated gene transcription—a possible mechanism. Br J Pharmacol 130: 1385–1393.
Budziszewska B, Jaworska-Feil L, Tetich M, Basta-Kaim A, Kubera M, Leśkiewicz M et al (2002). Effect of antidepressant drugs on the human corticotropin-releasing-hormone gene promoter activity in Neuro-2A cells. Pol J Pharmacol 54: 711–716.
Chatterjee D, Chakraborty M, Anderson GM (1992). Differentiation of Neuro-2a neuroblastoma cells by an antibody to GM3 ganglioside. Brain Res 583: 31–44.
Dąbrowski S, Kur J (1998). Cloning and expression in Escherichia coli of the recombinant his-tagged DNA polymerases from Pyrococcus furiosus and Pyrococcus woesei. Protein Express Purif 14: 131–138.
De Girolamo LA, Billett EE, Hargreaves AJ (2000). Effects of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine on differentiating mouse N2a neuroblastoma cells. J Neurochem 75: 133–140.
Delbende C, Tranchand Bunel D, Tarozzo G, Grino M, Oliver C, Mocaer E et al (1994). Effect of chronic treatment with the antidepressant tianeptine on the hypothalamo-pituitary-adrenal axis. Eur J Pharmacl 251: 245–251.
Fadda P, Pani L, Porcella A, Fratta W (1995). Chronic imipramine, L-sulpiride and mianserin decrease corticotropin releasing factor levels in the rat brain. Neurosci Lett 192: 121–123.
Glotzbach RK, Preskorn SH (1982). Brain concentration of tricyclic antidepressants: single-dose kinetics and relationship to plasma concentrations in chronically doses rats. Psychopharmacology 78: 25–27.
Groenink L, Dirks A, Verdouw PM, Schipholt M, Veening JG, van der Gugten J et al (2002). HPA axis dysregulation in mice overexpressing corticotropin releasing hormone. Biol Psychiatry 51: 875–881.
Guardiola-Diaz HM, Boswell C, Seasholtz AF (1994). The cAMP-responsive element in the corticotropin-releasing hormone gene mediates transcriptional regulation by depolarization. J Biol Chem 269: 14784–14791.
Guardiola-Diaz HM, Kolinske JS, Gates LH, Seasholtz AF (1996). Negative glucorticoid regulation of cyclic adenosine 3′,5′-monophosphate-stimulated corticotropin-releasing hormone-reporter expression in AtT-20 cells. Mol Endocrinol 10: 317–329.
Herman JP, Cullinan WE (1997). Neurocircuitry of stress: central control of the hypothalamo-pituitary-adrenocortical axis. Trends Neurosci 20: 78–84.
Holsboer F, Gerken A, Stalla GK, Muller OA (1985). ACTH, cortisol and corticosterone output after ovine corticotrophin-releasing factor challenge during depression and after recovery. Biol Psychiat 20: 276–286.
Hosoda K, Duman RS (1993). Regulation of beta 1-adrenergic receptor mRNA and ligand binding by antidepressant treatments and norepinephrine depletion in rat frontal cortex. J Neurochem 60: 1335–1343.
Hrdina PD, Dubas TC (1981). Brain distribution and kinetics of desipramine in the rat. Can J Physiol Pharmacol 59: 163–167.
Itoi K, Horiba N, Tozawa F, Sakai Y, Sakai K, Abe K et al (1996). Major role of 3′,5′-cyclic adenosine monophosphate-dependent protein kinase A pathway in corticotropin-releasing factor gene expression in the rat hypothalamus in vivo. Endocrinology 137: 2389–2396.
Jeffcoate WJ, Silverstone JT, Edwards CRW, Besser GM (1979). Psychiatric manifestations of Cushing's Syndrome: response to lowering of plasma cortisol. Q J Med 191: 465–472.
Kadota Y, Niiya A, Masaki R, Yamamoto A, Araki M, Taketani S (1997). A newly identified membrane protein localized exclusively in intracellular organelles of neurons. Brain Res Mol Brain Res 46: 265–273.
Karson CN, Newton JE, Mohanakrishnan P, Sprigg J, Komoroski RA (1992). Fluoxetine and trifluoperazine in human brain: a 19F-nuclear magnetic resonance spectroscopy study. Psychiatry Res 45: 95–104.
Klebe RJ, Ruddle FH (1969). Neuroblastoma: cell culture analysis of a differentiating stem cell system. J Cell Biol 43: 69.
Kole MH, Swan L, Fuchs E (2002). The antidepressant tianeptine persistently modulates glutamate receptor currents of the hippocampal CA3 commissural associational synapse in chronically stressed rats. Eur J Neurosci 16: 807–816.
Komoroski RA, Newton JE, Cardwell D, Sprigg J, Pearce J, Karson CN (1994). In vivo 19F spin relaxation and localized spectroscopy of fluoxetine in human brain. Magn Reson Med 31: 204–211.
Kornhuber J, Retz W, Riederer P (1995). Slow accumulation of psychotropic substances in the human brain. Relationship to therapeutic latency of neuroleptic and antidepressant drugs? J Neural Transm Suppl 46: 315–323.
Linkowski P, Mendlewicz J, LeClerc R, Brasseur M, Hubain P, Goldstein J et al (1985). The 24-hour profile of ACTH and cortisol in major depressive illness. J Clin Endocrinol Metab 61: 429–438.
Malkoski SP, Dorin RI (1999). Composite glucocorticoid regulation at a functionally defined negative glucocorticoid response element of the human corticotropin-releasing hormone gene. Mol Endocrinol 13: 1629–1644.
Manier DH, Shelton RC, Sulser F (2002). Noradrenergic antidepressants: does chronic treatment increase or decrease nuclear CREB-P? J Neural Transm 109: 91–99.
Miyake K, Fukuchi H, Kitaura T, Kimura M, Sarai K, Nakahara T (1990). Pharmacokinetics of amitriptyline and its demetiylated metabolite in serum and specific brain regions of rats after acute and chronic administration of amitriptyline. J Pharmaceut Sci 79: 288–291.
Murphy BEP (1997). Antiglucocorticoid therapies in major depression: a review. Psychoneuroendocrinology 22: S125–S132.
Nemeroff CB, Widerlov E, Bisette G, Walleus H, Karlsson I, Eklund K et al (1984). Elevated concentrations of CSF corticotropin releasing factor-like immunoreactivity in depressed patients. Science 226: 1342–1344.
Nestler EJ, McMahon A, Sabban EL, Tallman JF, Duman RS (1990). Chronic antidepressant administration decreases the expression of tyrosine hydroxylase in the rat locus coeruleus. Proc Natl Acad Sci USA 87: 7522–7526.
Nibuya M, Nestler EJ, Duman RS (1996). Chronic antidepressant administration increases the expression of cAMP response element binding protein (CREB) in rat hippocampus. J Neurosci 16: 2365–2372.
O'Brien D, Skelton KH, Owens MJ, Nemeroff CB (2001). Are CRF receptor antagonists potential antidepressants? Hum Psychopharmacol 16: 81–87.
Okuyama-Tamura M, Mikuni M, Kojima I (2003). Modulation of the human glucocorticoid receptor function by antidepressive compounds. Neurosci Lett 342: 206–210.
Pariante CM, Miller AH (2001). Glucocorticoid receptors in major depression: relevance to pathophysiology and treatment. Biol Psychiatry 49: 391–404.
Pariante CM, Pearce BD, Pisell TL, Owens MJ, Miller AH (1997). Steroid-independent translocation of the glucocorticoid receptor by the antidepressant desipramine. Mol Pharmacol 52: 571–581.
Post A, Crochemore C, Uhr M, Holsboer F, Behl C (2000). Differential induction of NF-kappaB activity and neural cell death by antidepressants in vitro. Eur J Neurosci 12: 4331–4337.
Raadsheer FC, Hoogendijk WJG, Stam FC, Tilders FJH, Swaab DF (1994). Increased numbers of corticotropin-releasing hormone expressing neurons in the hypothalamic paraventricular nucleus of depressed patients. Neuroendocrinology 460: 436–444.
Ratka A, Sutanto W, Bloemers M, De Kloet ER (1989). On the role of brain mineralocorticoid (type I) and glucocorticoid (type II) receptors in neuroendocrine regulation. Neuroendocrinology 50: 117–123.
Rosen LB, Majzoub JA, Adler GK (1992). Effects of glucocorticoid on corticotropin-releasing hormone gene regulation by second messenger pathways in NPLC and AtT-20 cells. Endocrinology 130: 2237–2244.
Rossby SP, Manier DH, Liang S, Nalepa I, Sulser F (1999). Pharmacological actions of the antidepressant venlafaxine beyond aminergic receptors. Int J Neuropsychopharmcol 2: 1–8.
Schwaninger M, Schofl C, Blume R, Rossig L, Knepel W (1995). Inhibition by antidepressant drugs of cyclic AMP response element-binding protein/cyclic AMP response element-directed gene transcription. Mol Pharmacol 47: 1112–1118.
Seasholtz AF, Thompson RC, Douglass JO (1988). Identification of a cyclic adenosine monophosphate-responsive element in the rat corticotropin-releasing hormone gene. Mol Endocrinol 2: 1311–1319.
Shibahara S, Morimoto Y, Furutani Y, Notake M, Takahashi H, Shimizu S et al (1983). Isolation and sequence analysis of the human corticotropin-releasing factor precursor gene. EMBO J 2: 775–779.
Skaper SD, Adelson GL, Seegmiller JE (1976). Metabolism of biogenic amines in neuroblastoma and glioma cells in culture. J Neurochem 27: 1065–1070.
Spengler D, Rupprecht R, Van LP, Holsboer F (1992). Identification and characterization of a 3′,5′-cyclic adenosine monophosphate-responsive element in the human corticotropin-releasing hormone gene promoter. Mol Endocrinol 6: 1931–1941.
Stout SC, Owens MJ, Nemeroff CB (2002). Regulation of corticotropin-releasing factor neuronal systems and hypothalamic–pituitary–adrenal axis activity by stress and chronic antidepressant treatment. J Pharmacol Exp Ther 300: 1085–1092.
Tamura T, Morinobu S, Okamoto Y, Kagaya A, Yamawaki S (2002). The effects of antidepressant drug treatments on activator protein-1 binding activity in the rat brain. Prog Neuropsychopharmacol Biol Psychiatry 26: 375–381.
Vamvakopoulos NC, Chrousos GP (1994). Hormonal regulation of human corticotropin-releasing hormone gene expression: implications for the stress response and immune/inflammatory reaction. Endocrine Rev 15: 409–420.
Van LP, Spengler DH, Holsboer F (1990). Glucocorticoid repression of 3′,5′-cyclic-adenosine monophosphate-dependent human corticotropin-releasing-hormone gene promoter activity in a transfected mouse anterior pituitary cell line. Endocrinology 127: 1412–1418.
Watanabe Y, Gould E, Daniels DC, Cameron H, McEwen BS (1992). Tianeptine attenuates stress-induced morphological changes in the hippocampus. Eur J Pharmacol 222: 157–162.
Woods MD, Shipston MJ, Mullens EL, Antoni FA (1992). Pituitary corticotrope tumor (AtT20) cells as a model system for the study of early inhibition by glucocorticoids. Endocrinology 131: 2873–2880.
Zobel AW, Nickel T, Kunzel HE, Ackl N, Sonntag A, Ising M et al (2000). Effects of the high-affinity corticotropin-releasing hormone receptor 1 antagonist R121919 in major depression: the first 20 patients treated. J Psychiatr Res 34: 171–181.
Acknowledgements
We are grateful to Dr Anna Brillowska-Dąbrowska and Dr Sławomir Dąbrowski for hCRHCAT and hCMVCAT plasmid synthesis. We thank Ms B Korzeniak for her skillful technical assistance. This study was supported by the Grant 6 P05A 069 20 from the State Committee for Scientific Research, Warszawa, Poland.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Budziszewska, B., Jaworska-Feil, L., Tetich, M. et al. Regulation of the Human Corticotropin-Releasing-Hormone Gene Promoter Activity by Antidepressant Drugs in Neuro-2A and AtT-20 Cells. Neuropsychopharmacol 29, 785–794 (2004). https://doi.org/10.1038/sj.npp.1300379
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.npp.1300379
Keywords
This article is cited by
-
Genome-wide association study of antidepressant response: involvement of the inorganic cation transmembrane transporter activity pathway
BMC Psychiatry (2016)
-
The Antidepressant Desipramine Requires the ABCB1 (Mdr1)-Type p-Glycoprotein to Upregulate the Glucocorticoid Receptor in Mice
Neuropsychopharmacology (2007)
-
Antipsychotic Drugs Inhibit the Human Corticotropin-Releasing-Hormone Gene Promoter Activity in Neuro-2A Cells—an Involvement of Protein Kinases
Neuropsychopharmacology (2006)
-
Increased mortality in depressive disorders: A review
Current Psychiatry Reports (2004)