Abstract
Nucleotide excision repair (NER) pathway plays critical roles in repairing DNA disorders caused by platinum. To comprehensively understand the association between variants of NER and clinical outcomes of platinum-based chemotherapy, 173 SNPs in 27 genes were selected to evaluate association with toxicities and efficiency in 1004 patients with advanced non-small cell lung cancer. The results showed that consecutive significant signals were observed in XPA, RPA1, POLD1, POLD3. Further subgroup analysis showed that GTF2H4 presented consecutive significant signals in clinical benefit among adenocarcimoma. In squamous cell carcinoma, rs4150558, rs2290280, rs8067195 were significantly associated with anemia, rs3786136 was significantly related to thrombocytopenia, ERCC5 presented consecutive significant signals in response rate. In patients receiving TP regimen, significant association presented in neutropenia, thrombocytopenia and gastrointestinal toxicity. Association with anemia and neutropenia were found in GP regimen. rs4150558 showed significant association with anemia in NP regimen. In patients > 58, ERCC5 showed consecutive significant signals in gastrointestinal toxicity. Survival analysis showed SNPs in POLD2, XPA, ERCC6 and POLE were significantly associated with progression free survival, SNPs in GTF2H4, ERCC6, GTF2HA, MAT1, POLD1 were significantly associated with overall survival. This study suggests SNPs in NER pathway could be potential predictors for clinical outcomes of platinum-based chemotherapy among NSCLC.
Similar content being viewed by others
Introduction
Lung cancer is one of the most common cancer and the leading cause of cancer-related death worldwide1. Despite the improvements of diagnosis and treatment, the prognosis of lung cancer is still poor, and the 5-year-survival rates vary from 4–17% depending on stage and regional differences2. Non-small cell lung cancer (NSCLC) accounts for about 80% of primary lung cancer and most patients suffered advanced disease at the time of diagnosis. Two major types of NSCLC are adenocarcimoma (AC) and squamous cell carcinoma (SCC)3.
Platinum is an effective antitumor agent and platinum-based chemotherapy is widely used in various cancer treatment4. The most commonly used platinum containing agents clinically are cisplatin, carboplatin, oxaliplatin. Cisplatin is first discovered and very commonly used to treat many tumors, including lung cancer5,6. The anti-tumor mechanism of platinum compounds is to disorder the DNA replication and induce cell death7,8. The most common adduct formed by platinum is intra-strand cross. Cisplatin and carboplatin have the same cross-link, which is 1,2-intrastrand cross links between adjacent purine bases, and oxaliplatin presents a structurally distinct adduct containing a bulky 1,2-diaminocyclohexane group9. If the adducts caused by platinum could not be repaired, the disordered DNA could inhibit DNA replication progression, and drive cells into apoptosis10.
The damage caused by platinum is recognized and repaired mainly through nucleotide excision repair (NER) pathway11,12. DNA damage is repaired by NER via four processes: DNA damage recognition, DNA unwinding, DNA incision, and DNA resynthesis and ligation10,13. Many genes involve in these processes. XPC, ERCC6, and ERCC8 play important roles in DNA damage recognition, ERCC2, ERCC3, XPA, and RPA1 participate in DNA unwinding, ERCC1, ERCC4, ERCC5 are responsible for DNA incision10. More and more evidences showed that NER was an important mediator of tumor sensitivity to platinum. For example, low expression level of XPA and ERCC1 increased patients′ sensitivity to cisplatin14,15, while high level of ERCC1 was significantly associated with cisplatin resistance. The expression level of ERCC1 was considered as a potential biomarker for response to cisplatin-based chemotherapy16,17. Some studies showed that single nucleotide polymorphisms (SNPs) in NER pathway were also significantly associated with various cancer risk and prognosis, especially the response to platinum-based chemotherapy18,19,20. Some reviews pointed out that there was huge potential clinical value in using mRNA or protein levels of NER genes to predict the response to cisplatin-based chemotherapy for NSCLCs8,10, however, the results of studies which investigated the association between SNPs of NER and clinical outcomes of platinum-based treatment are not consistent. In order to fully evaluate the potential clinical value of the SNPs of NER pathway in predicting clinical outcomes of platinum-based chemotherapy for NSCLCs, 1004 Chinese patients with advanced NSCLC who received only platinum-based treatment were enrolled in this study. 173 SNPs located in 27 genes of NER pathway were selected to assess the association between these SNPs and clinical outcomes of platinum-based chemotherapy, including gastrointestinal toxicity, neutropenia, anemia, thrombocytopenia, clinical benefit, response rate, overall survival (OS), and progression-free survival (PFS).
Results
Characteristics of patients and clinical outcomes
In order to investigate the association between polymorphisms of NER pathway and clinical outcomes of platinum-based chemotherapy, 1004 patients with advanced NSCLC who received only first-line platinum-based chemotherapy were enrolled in this study. The details of patient characteristics and clinical outcomes were listed in Table 1. The median age of cohort was 58 (ranged from 26 to 82). The patients who were more than 58-year-old accounted for 48.4%, and the ones who were less than or equal to 58-year-old accounted for 51.6%. Most patients were male (70.3%). The percentage of patients with ECOG PS 0–1 was 91.3%. 42.5% of the patients were non-smoker. All patients recruited presented advanced NSCLC, and most of which were stage IV (62.6%). Adenocarcinoma was the most common histological type, which accounted for 57.5%. Platinum-navelbine (NP) (31.5%), platinum-gemcitabine (GP) (23.8%), platinum-paclitaxel (TP) (31.1%), platinum-docetaxel (DP) (8.7%) were the four mainly used chemotherapy regimens in this study. The responses of platinum-based chemotherapy were classified into 4 categories in terms of complete response (CR), partial response (PR), stable disease (SD), and progressive disease (PD) according to Response Evaluation Criteria in Solid Tumors (version 1.0)21. Clinical benefit was defined as patients with CR, PR or SD. Response rate contains CR and PR. The response rate was 18.2%, and clinical benefit was 80.7%. The median PFS was 9.1 months and the median OS was 19.3 months. In the toxicity analysis, gastrointestinal toxicity and hematological toxicities including anemia, thrombocytopenia, and neutropenia were collected. 8.3% of patients presented severe gastrointestinal toxicity, 3.1% of patients presented severe anemia, 12.3% of patients presented severe neutropenia, and 3.6% of patients presented severe thrombocytopenia.
Association between the polymorphisms of NER pathway and efficiency of platinum-based chemotherapy
To investigate the association between polymorphisms of NER pathway and the efficiency of platinum-based chemotherapy, clinical benefit and response rate were introduced in this study to evaluate the efficacy of platinum-based chemotherapy. There were many polymorphisms presented significant association with clinical benefit and/or response rate of platinum-based chemotherapy (P < 0.05), however, after Bonferroni correction, no significant results were remained (P < 2.89 × 10−4 (0.05/173)) (Fig. 1A). rs3176721 located in XPA showed the most significant signal in clinical benefit analysis (χ2 test P = 0.003; OR = 1.74, 95%CI:1.25–2.44, P = 0.001).
Association analysis between polymorphisms of NER pathway and outcomes of platinum-based chemotherapy in lung cancer. Red line means the significance level after strict Bonferroni correction (P < 2.89 × 10−4 ((0.05/173)), black line means the significance level of 0.05. (A) association analysis in all patients; (B) association analysis in subgroup of adenocarcinoma; (C) association analysis in subgroup of squamous cell carcinoma; (D) association analysis in subgroup of paclitaxel combined with cisplatin regimen; (E) association analysis in subgroup of gemcitabine combined with cisplatin regimen; (F) association analysis in subgroup of navelbine combined with cisplatin regimen; (G) association analysis in subgroup of age ≤ 58; (H) association analysis in subgroup of age > 58. The genes analyzed in this study is as follow: 1, XPC; 2, RAD23B; 3, ERCC2; 4, GTF2H1; 5, XPA; 6, ERCC5; 7, ERCC1; 8, ERCC4; 9, ERCC8; 10, ERCC6; 11, DDB2; 12, LIG1; 13, CDK7; 14, CCNH; 15, MNAT1; 16, RPA1; 17, RPA2; 18, RFC1; 19, RFC2; 20, POLD1; 21, POLD2; 22, POLD3; 23, POLD4; 24, POLE; 25, POLE2; 26, GTF2H3; 27, GTF2H4.
Subgroup analyses in different histological types showed that 6 SNPs of ERCC5 presented consecutive significant signals in response rate in SCC, and rs2296147 showed the most significant result (χ2 test P = 4.13 × 10−4; OR = 0.34, 95%CI:0.20–0.59, P = 9.70 × 10−5) (Fig. 1C). 4 SNPs located in GTF2H4 (also known as P52) presented consecutive significant signals in clinical benefit in AC and the most significant locus was rs3218804 (χ2 test P = 0.001; OR = 2.29, 95%CI:1.43–3.66, P = 0.001), although no SNPs reached the significant level of Bonferroni correction (Fig. 1B) (Table 2).
Subgroup analysis among patients receiving different chemotherapy regimens showed that no polymorphisms could achieve the significant level of Bonferroni correction. However, in subgroup of patients receiving NP regimen (Fig. 1F), ERCC5 and DDB2 presented consecutive significant signals in clinical benefit, and the most significant signals were rs2228959 (χ2 test P = 0.003; OR = 2.03, 95%CI:1.04–3.94, P = 0.037) in ERCC5 and rs2306353 (χ2 test P = 0.001; OR = 0.49, 95%CI:0.29–0.82, P = 0.007) in DDB2. ERCC2 showed consecutive significant signals in response rate, and the most significant SNP was rs238406 (χ2 test P = 0.003; OR = 0.64, 95%CI:0.43–0.95, P = 0.025). In subgroup of patients treated with TP regimen (Fig. 1D), ERCC5 and ERCC1 showed consecutive significant signals in response rate, and the most significant SNP was rs873601 (χ2 test P = 0.005; OR = 2.48, 95%CI:1.30–4.75, P = 0.006) in ERCC5, rs3212961 (χ2 test P = 0.002; OR = 0.54, 95%CI:0.34–0.86, P = 0.009) in ERCC1 (Table 2). No significant association between polymorphisms of NER pathway and clinical benefit or response rate of platinum-based chemotherapy was found in patients receiving GP regimen (Fig. 1E).
Association between polymorphisms of NER pathway and the toxicities of platinum-based chemotherapy
Gastrointestinal toxicity and hematological toxicities including anemia, thrombocytopenia, and neutropenia were collected to investigate the association between SNPs of NER pathway and the toxicities of platinum-based chemotherapy. The results showed that GTF2H1/P62 and DDB2 presented consecutive significant signals on anemia. RPA1 and POLD1 presented consecutive significant signals on thrombocytopenia. POLD3 presented consecutive significant signals on neutropenia (Fig. 1A). However, no SNPs satisfied the significant level of Bonferroni correction (P < 2.89 × 10−4).
Subgroup analyses in different histological types showed that rs3786136 in RPA1 were significantly associated with thrombocytopenia in SCC (χ2 test P = 3.13 × 10−5; OR = 4.71, 95%CI:1.10–20.12, P = 0.037) (Fig. 1C) after Bonferroni correction. rs4150558 (χ2 test P = 1.61 × 10−6; OR = 23.45, 95%CI:2.64–208.13, P = 0.005) in GTF2H1, rs2290280 (χ2 test P = 2.86 × 10−6; OR = 28.53, 95%CI:1.69–481.13, P = 0.020) in CCNH, rs8067195 (χ2 test P = 1.01 × 10−5; OR = 6.93, 95%CI:1.44–33.49, P = 0.016) and rs6416887 (χ2 test P = 3.07 × 10−5; OR = 6.55, 95%CI:1.32–32.44, P = 0.021) in RPA1 were significantly related to anemia in SCC (Fig. 1C) (Table 2).
Subgroup analyses among patients receiving different chemotherapy regimens showed that in subgroup of patients receiving TP regimen (Fig. 1D), rs4253002 in ERCC6 was significantly associated with gastrointestinal toxicity (χ2 test P = 1.26 × 10−4; OR = 7.81, 95%CI:2.27–26.88, P = 0.001). rs4151405 in MNAT1(P = 4.58 × 10−5) and rs17584703 in RFC1 (P = 9.72 × 10−7) showed significantly different distribution in thrombocytopenia, however, multiple logistic regression analysis showed that there were no significant association between the 2 SNPs and thrombocytopenia. rs1726801 (χ2 test P = 3.27 × 10−5; OR = 3.03, 95%CI:1.59–5.77, P = 0.001), rs1673041 (χ2 test P = 3.27 × 10−5; OR = 3.46, 95%CI:1.97–6.09, P = 1.70 × 10−5) and rs3219341 (χ2 test P = 3.09 × 10−5; OR = 3.03, 95%CI:1.59–5.75, P = 0.001) in PLOD1 were significantly associated with neutropenia (Table 2). In subgroup of patients receiving GP regimen (Fig. 1E), rs4253212 (χ2 test P = 4.92 × 10−5; OR = 3.31, 95%CI:1.26–8.72, P = 0.015) in ERCC6 was significantly associated with neutropenia. rs1799793 (χ2 test P = 2.71 × 10−5; OR = 7.91, 95%CI:2.02–30.96, P = 0.003) in ERCC2 was significantly associated with anemia. rs20580 (χ2 test P = 0.001; OR = 3.21, 95%CI:1.53–6.74, P = 0.002) in LIG1 was significantly associated with gastrointestinal toxicity (Table 2). We also found rs4150558 (χ2 test P = 1.24 × 10–5; OR = 4.39, 95%CI:1.37–14.08, P = 0.013) in GTF2H1 were significantly associated with anemia in patients receiving NP regimen (Fig. 1F) (Table 2).
Subgroup analyses in the age of patients ≤ 58 (Fig. 1G) showed that DDB2 and RPA1 presented consecutive significant signals on neutropenia. rs326222 (χ2 test P = 7.43 × 10−5; OR = 2.07, 95%CI:1.32–3.23, P = 0.001) in DDB2 remained significant association with neutropenia after Bonferroni correction (Table 2). In the subgroup of patients who were over 58-year-old (Fig. 1H), ERCC5 showed consecutive significant signals in gastrointestinal toxicity, and 3 SNPs including rs4150339 (χ2 test P = 2.10 × 10−7; OR = 2.53, 95%CI:1.23–5.22, P = 0.012), rs2296147 (χ2 test P = 3.88 × 10−5; OR = 2.10, 95%CI:1.21–3.64, P = 0.008) and rs4150360 (χ2 test P = 1.05 × 10−4; OR = 3.07, 95%CI:1.70–5.55, P = 2.12 × 10−4) remained significant association with gastrointestinal toxicity after Bonferroni correction (Table 2).
Association between polymorphisms of NER and survival of platinum-based chemotherapy
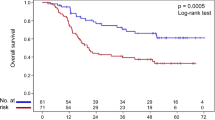
Survival analysis was performed to assess the association between the polymorphisms of NER and PFS or OS. The results showed that 5 SNPs were associated with PFS, and all these SNPs decreased the risk of disease progression (Table 3, Fig. 2A–E). rs3757843 (Log-rank P = 0.004; HR = 0.78, 95%CI:0.65–0.93, P = 0.005) in POLD2, rs3176658 (Log-rank P = 0.007; HR = 0.81, 95%CI:0.68–0.96, P = 0.015) in XPA, rs11609456 (Log-rank P = 0.002; HR = 0.76, 95%CI:0.62–0.94, P = 0.010) and rs5744751 (Log-rank P = 0.003; OR = 0.77, 95%CI:0.62–0.94, P = 0.011) in POLE presented significant association in dominant model. rs12571445 (Log-rank P = 0.020; OR = 0.13, 95%CI:0.02–0.93, P = 0.042) in ERCC6 presented significant association when assuming recessive model. In the analysis of OS (Table 4, Fig. 2F–J), rs3130780 (Log-rank P = 0.003; HR = 13.65, 95%CI:1.88–99.37, P = 0.010) in GTF2H4, rs4150667 (Log-rank P = 0.017; HR = 1.36, 95%CI:1.06–1.75, P = 0.015) in GTF2H1, and rs2546551 (Log-rank P = 0.002; HR = 1.84, 95%CI:1.15–2.94, P = 0.011) in POLD1 increased the risk of death in recessive model. rs4151374 (Log-rank P = 0.036; HR = 0.86, 95%CI:0.75–0.99, P = 0.049) in MAT1 played a significantly protective role in dominant model. rs2281793 (Log-rank P = 0.007; HR = 0.70, 95%CI:0.53–0.91, P = 0.009) in ERCC6 could prolong patients’ OS when assuming recessive model.
PFS and OS curves of significant polymorphisms of NER pathway. Best models were used in the analysis. (A–E) showed the results of PFS, and (F–J) showed the results of OS. (A) rs3757843; (B) rs3176658; (C) rs12571445; (D) rs11609456; (E) rs5744751; (F) rs3130780; (G) rs2281793; (H) rs4150667; (I) rs4151374; (J) rs2546551.
Discussion
NER pathway is important in DNA damage repair, especially in repairing the distortion of DNA helical structure22. Many genes involved in lesion recognition, DNA unwinding, incision of the DNA around lesion, and finally DNA resynthesis and ligation13. Platinum-based chemotherapy is one of the most effective treatments for lung cancer. The mechanism of platinum in cancer treatment is to form intra and inter-strand crosslinks, which could distort the DNA helix, inhibit DNA replication and cause cancer cells apoptosis5. NER pathway is the main damage repair system involved in platinum-caused DNA distortion4. Many studies focused on the relationship between the expression level of NER-related genes and efficacy of platinum-based treatment for cancer. The status of ERCC1 protein expression was reported as a predictive marker for outcomes of platinum-based chemotherapy in lung cancer17. Some studies also pointed out those SNPs in some members of NER pathway showed significant association with clinical outcomes of platinum-based chemotherapy. The polymorphisms of XPD were significantly associated with not only efficiency but also severe toxicity of platinum-based chemotherapy in lung cancer23,24. Other members of NER pathway, such as XPA, ERCC5, and ERCC2, were related to the response of platinum-based chemotherapy in lung cancer15,25,26. In order to comprehensively assess the association between polymorphisms of NER pathway and clinical outcomes of platinum-based chemotherapy, a total of 173 SNPs located in 27 genes were investigated in this study to evaluate their association with gastrointestinal toxicity, neutropenia, anemia, thrombocytopenia, clinical benefit, response rate, overall survival (OS), and progression-free survival (PFS).
Our results showed that variants in NER pathway were significantly associated with clinical outcomes of platinum-based chemotherapy. Polymorphisms in XPA, DDB2 and GTF2H4 were significantly associated with clinical benefit. Polymorphisms in ERCC2, ERCC5 were significantly associated with response rate. Polymorphisms in GTF2H1, ERCC2 and RPA1 showed significant association with anemia. Polymorphisms in RPA1 showed significant association with thrombocytopenia. Polymorphisms in ERCC2, ERCC6, DDB2, RPA1, POLD1 and POLD3 presented significant association with neutropenia. Polymorphisms in POLD2, XPA, ERCC6, POLE presented significant association with PFS. Polymorphisms in GTF2H4, ERCC6, GTF2H1, MAT1 and POLD1 presented significant association with OS.
XPA encodes a zinc-finger DNA-binding protein, and plays an important role of damage recognition in NER pathway27. Genetic variants in XPA were significantly associated with lung cancer risk28. Knockdown the expression of XPA could sensitize NSCLC-derived cell lines to cisplatin29. Our results showed that rs3176721 in XPA was significantly associated with clinical benefit in all patients, as well as in AC subgroup. rs3176658 in XPA was significantly associated with PFS, and the A allele could significantly decrease the risk of disease progression.
DDB2 is a component of DDB which is the damage-specific DNA-binding heterodimeric complex30. SNPs in DDB2 were significantly associated with the risk of lung cancer31. A recent GWAS analysis showed that rs747650 in DDB2 was a new susceptibility locus of severe acne32. Overexpression of DDB2 could sensitize the cancer cells to cisplatin treatment which indicated that DDB2 may play important role in platinum-based chemotherapy33. In our study, we found that rs2306353 significantly associated with clinical benefit in patients receiving NP regimen, and rs326222 in DDB2 were significantly risk factor for neutropenia in subgroup of patients younger than 58 years old.
GTF2H4 (also known as P52) encodes a subunit of transcription factor II H (TFIIH), and is known to be involved in nucleotide excision repair34. In a recent study of a large-scale analysis of six published GWAS datasets pointed out that rs114596632 in GTF2H4 was significantly associated with lung cancer risk35, rs2074508 in GTF2H4 was significantly associated with smoking-related lung cancer36. In the current study, GTF2H4 presented consecutive significant signals in clinical benefit among AC patients. rs3130780 in GTF2H4 was significantly associated with OS, and AA genotype could significantly increase risk of death.
ERCC5 plays important roles in DNA incision in NER pathway. ERCC5 is a well-known gene which has great impact on cancer. Our study showed that ERCC5 presented consecutive significant signals not only in response rate in SCC, but also in gastrointestinal toxicity among patients > 58 years old. rs2296147 was the most significant SNP which associated with response rate. It was reported that rs2296147 was not only associated with cancer risk, but also related to prognosis of cancer37. There were also many studies showed that rs2296147 was associated with prognosis of advanced non-small cell lung cancer treated with platinum-based chemotherapy, and could predict the clinical outcomes of platinum-based chemotherapy38,39,40,41. rs2296147 is located in the promoter of ERCC5. The transcription repressor of SNAI1 is predicted to bind to the sequence around rs2296147, which indicating that rs2296147 may take part in negative regulating the expression of ERCC5.
RPA1 is an important subunit of RPA which is a major eukaryotic single-strand DNA-binding protein complex, and essential for DNA repair, DNA replication, DNA recombination, telomere maintenance, activation of DNA damage checkpoints and the maintenance of genomic integrity42. RPA1 is also reported as a part of the replication fork protection complex43. Previous studies showed that RPA1 played important roles in Pt-DNA repair44, and expression level of RPA1 could be used to predict prognosis of cancer45. However, no studies focused on the relationship between RPA1 and the hematological toxicities of platinum-based chemotherapy. In this study, we found that polymorphisms in RPA1 presented significant association with all 3 hematological toxicities. rs12727 and rs3786136 showed significant association with thrombocytopenia, rs8067195 and rs6416887 showed significant association with anemia, rs12150513 showed significant association with neutropenia. rs12727 is located in the 3′UTR of RPA1, and the sequence around it is the potential target of miR-345-3p, miR-6732-3p and miR-6771-3p. RPA1 is also a target of PTEN function in fork protection to maintain genome stability46.
ERCC6 can recognize DNA damage and recruit NER repair factors to the DNA damage site. Polymorphisms in ERCC6 showed significant association with the risk and prognosis of lung cancer47. Previous study showed that no statistically significant association was found between the platinum-related toxicities and SNPs of ERCC6 or, CCNH 48. In our study, we found that rs4253002 in ERCC6 showed significant association with gastrointestinal toxicity in the patients receiving TP regimen, and rs4253212 in ERCC6 showed significant association with neutropenia in the patients receiving GP regimen. We also found rs2290280 in CCNH was significantly associated with anemia in SCC subgroup. In survival analysis, rs12571445 in ERCC6 showed significant association with PFS, and rs2281793 in ERCC6 showed significant association with OS. Our results suggested that both ERCC6 and CCNH might involve in regulating clinical outcomes of platinum-based chemotherapy.
DNA polymerase δ is conserved from humans to yeast, and performs important functions in DNA replication and repair processes. The Polδ complex was comprised of four subunits (p125, p66, p50 and p12) which encoded by POLD1, POLD3, POLD2 and POLD4 49. Polymorphisms and mutations in POLD1 and POLD3 were reported to be associated with cancer risk50,51. Overexpression of POLD1 was associated with platinum resistance in a long-term survivor of mesothelioma52. In this study, POLD1 and POLD3 showed significant association with neutropenia. rs1726801, rs1673041 and rs3219341 in POLD1 showed significant association with neutropenia in patients receiving TP regimen. rs10857 and rs6592576 in POLD3 showed significant association with neutropenia in all patients. rs3757843 in POLD2 showed significant association with PFS, and rs2546551 in POLD1 showed significant association with OS.
We also found that rs11609456 and rs5744751 in POLE showed significant association with PFS, rs4151374 in MAT1 and rs4150667 in GTF2H1 showed significant association with OS. rs4150558 in GTF2H1 was significantly associated with anemia in all patients, the same effect was also observed in not only SCC but also subgroup of patients receiving NP regimen. Our results showed that some of the significant signals of χ2 test were absent in multiple logistic regression analysis, especially in subgroup analysis. For example, rs12727 in RPA1 showed in significantly different distribution in thrombocytopenia in AC subgroup, rs4151405 in MNAT1 and rs17584703 in RFC1 showed significantly different distribution in thrombocytopenia in patients receiving TP regimen, however, multiple logistic regression analysis showed no significant association. This might be because that the number of patients were few in some subgroups, resulting in the distribution of genotypes disequilibrium and significant signals of χ2 test. However, P value for trend as well as OR and 95%CI were used in multiple logistic regression analysis, which reveal the real relationship or association between clinical outcomes and polymorphisms.
In the current study, subgroups analysis of chemotherapy regimen was carried out to investigate other drugs affect the results of association analysis of platinum. We found that different genes were associated with different outcomes in different subgroups, which suggested that other drugs effect might have impact on clinical outcomes of platinum-based treatment and subgroup analysis was important in platinum-related pharmacogenetics studies. In survival analysis, some significant signals were only presented in heterozygote, but disappeared in mutant homozygote. This phenomenon was termed “heterozygote advantage”. Many other studies showed the similar results. For example, there was a clear association between heterozygosity at the TIRAP S180L locus and protection against multiple infectious diseases53. In breast cancer that the heterozygous genotype of 5′ UTR -26 G > A polymorphism located in BRCA2 was found to be protective effect in cancer risk. Our results also showed that heterozygous genotype was significantly associated with good prognosis54. In some subgroups of survival analysis, especially in recessive model, such as rs12571445 (ERCC6) in PFS analysis, and rs3130780 (GTF2H4) and rs2546551 (POLD1) in OS analysis, the sample size of homozygous mutation is too small to get reliable results, and more samples are needed to confirm the results.
Summary, 173 SNPs located in 27 genes of NER pathway were investigated in this study to assess the association with clinical outcomes of platinum-based chemotherapy for advanced NSCLC. SNPs in ERCC2 (rs1799793), ERCC5 (rs4150339, rs2296147, rs4150360, rs4771436), ERCC6 (rs4253002, rs4253212, rs12571445, rs2281793), XPA (rs3176721, rs3176658), GTF2H1 (rs4150558, rs4150667), GTF2H4 (rs3218804, rs3130780), DDB2 (rs326222), RPA1 (rs12727, rs8067195, rs6416887, rs3786136, rs12150513), POLD1 (rs3219281, rs3219341, rs1726801, rs1673041, rs2546551), POLD2 (rs3757843), POLD3 (rs10857, rs6592576), POLE (rs11609456, rs5744751) and MAT1 (rs4151374) showed significant association with toxicities and efficiency of platinum-based chemotherapy in different subgroups. Due to the low incidence of severe toxicity, statistics power is not sufficient in some groups, validation assay and functional investigation is needed in future study.
Methods
Study population
1004 patients recruited in current study were histopathologically diagnosed stage IIIA-IV NSCLC patients in Shanghai, China. Each patient was informed consent before enrolled. The criteria for recruitment were defined as below: (1) the patients enrolled in this study was over 18 years old; (2) the patients were newly diagnosed, and only received platinum-based chemotherapy. Any patient with surgery, radiotherapy, concurrent chemoradiotherapy or previous chemotherapy was excluded; (3) the performance status was between 0 and 2; (4) there were no other malignancy in the past 5 years; (5) no cardiac arrhythmias, no active congestive heart failure, and no uncontrolled clinical infections; (6) the absolute neutrophil count ≥ 1.5 × 109 cells/L, platelets ≥ 100 × 109cells/L, creatinine clearance ≥ 60 mL/min, serum creatinine ≤ 1.5 × upper limit normal, alanine and aspartate aminotransferase ≤ 1.5 × upper limit normal. All the methods mentioned in the protocol were carried out in accordance with the institutional guidelines and approved by the Ethical Review Committee of Fudan University, and informed consent was obtained from all patients before samples collection.
Clinical outcomes including toxicities, responses and survival were evaluated in the current study. The responses to platinum-based chemotherapy were assessed after two cycles of treatment, and the responses were classified into 4 categories in terms of complete response (CR), partial response (PR), stable disease (SD), and progressive disease (PD) according to response evaluation criteria in solid tumors (version 1.0)21. Clinical benefit was defined as patients with CR, PR or SD. Response rate contains CR and PR. Gastrointestinal toxicity and hematologic toxicities including neutropenia, anemia, and thrombocytopenia, were collected and evaluated twice a week according to the Common Terminology Criteria for Adverse Events V3.0 (CTCAE 3.0). Grade 3 or 4 toxicities were defined as severe adverse effects. Grade 5 toxicity, also known as death, was not observed in this study. Progression-free survival (PFS) and overall survival (OS) were assessed in the survival analysis. PFS was calculated from the date of first cycle of platinum-based chemotherapy to the date of PD, death, or the last follow-up. OS was calculated from the date of first cycle of platinum-based chemotherapy to the date of death or the last follow-up. The survival data was collected from follow-up calls, and the Social Security Death Index and inpatient and outpatient clinical medical records.
SNPs selection and genotyping
Base on the genotype data of Han Chinese in Beijing (CHB) from phase II Hapmap SNP database, 173 SNPs of 27 genes involved in NER pathway were selected using the strategies of tag-SNPs and functional SNPs by Haplowview 4.1 (http://www.broadinstitute.org/haploview) with the criteria of minor allele frequency ≥ 0.05 and correlation coefficient ≥ 0.8. The detail information was listed in Supplementary Table 1.
Human genomic DNA was extracted from blood samples using Qiagen Blood Kit (Qiagen, CA). All SNPs were genotyped using iSelect HD BeadChip (Illumina, San Diego, Calif). The results of random duplicate assays were consistent. Following the criteria of SNP genotyping call rate > 0.95, MAF > 0.01, GenCall score > 0.2, all 173 SNPs located in 27 genes (detailed in supplementary Table 1) were included in final analysis.
Statistical analysis
Demographic and clinical factors were test against clinical outcomes by chi-square tests or log-rank test. Factors that had P-value < 0.05 were regarded as covariates (Supplementary Table 2, Supplementary Table 3). The Chi-square test was used to assess whether SNPs’ genotypes were significantly different in the distribution of clinical outcomes. Bonferroni correction was performed by multiplying the number of all SNPs tested in the study to control for multiple comparisons. Significant SNPs from Chi-square were included in multiple logistic regression adjusted for covariates to estimate their association with clinical outcomes by odds ratio (OR) and confidence interval (CI). Log-rank test was used to compare the survival curve between patients’ groups. Cox proportional hazards regression adjusted for covariates was performed to evaluate the association between survival and significant polymorphisms SNPs from log-rank test by hazard ratios (HRs) with 95% CIs in additive, dominant, or recessive model. All P-values presented were two-sided, and a level of P < 0.05 was considered statistically significant. SPSS software (SPSS, Chicago, IL) and PLINK v1.07 were used for statistical analyses in this study.
References
Siegel, R., Ward, E., Brawley, O. & Jemal, A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA: a cancer journal for clinicians 61, 212–236 (2011).
Hirsch, F. R. et al. Lung cancer: current therapies and new targeted treatments. Lancet 389, 299–311 (2017).
Tang, E. R., Schreiner, A. M. & Pua, B. B. Advances in lung adenocarcinoma classification: a summary of the new international multidisciplinary classification system (IASLC/ATS/ERS). Journal of thoracic disease 6, S489–501 (2014).
Rosell, R. et al. Nucleotide excision repair pathways involved in Cisplatin resistance in non-small-cell lung cancer. Cancer control: journal of the Moffitt Cancer Center 10, 297–305 (2003).
Arora, S., Kothandapani, A., Tillison, K., Kalman-Maltese, V. & Patrick, S. M. Downregulation of XPF-ERCC1 enhances cisplatin efficacy in cancer cells. DNA repair 9, 745–753 (2010).
Rosenberg, B., VanCamp, L., Trosko, J. E. & Mansour, V. H. Platinum compounds: a new class of potent antitumour agents. Nature 222, 385–386 (1969).
Zamble, D. B. & Lippard, S. J. Cisplatin and DNA repair in cancer chemotherapy. Trends in biochemical sciences 20, 435–439 (1995).
O’Grady, S. et al. The role of DNA repair pathways in cisplatin resistant lung cancer. Cancer treatment reviews 40, 1161–1170 (2014).
Dasari, S. & Tchounwou, P. B. Cisplatin in cancer therapy: molecular mechanisms of action. European journal of pharmacology 740, 364–378 (2014).
Bowden, N. A. Nucleotide excision repair: why is it not used to predict response to platinum-based chemotherapy? Cancer letters 346, 163–171 (2014).
Wang, G., Dombkowski, A., Chuang, L. & Xu, X. X. The involvement of XPC protein in the cisplatin DNA damaging treatment-mediated cellular response. Cell research 14, 303–314 (2004).
Yen, L. et al. Regulation of cellular response to cisplatin-induced DNA damage and DNA repair in cells overexpressing p185(erbB-2) is dependent on the ras signaling pathway. Oncogene 14, 1827–1835 (1997).
Marini, F. et al. DNA nucleotide excision repair-dependent signaling to checkpoint activation. Proceedings of the National Academy of Sciences of the United States of America 103, 17325–17330 (2006).
Fu, X. et al. High expression of XPA confers poor prognosis for nasopharyngeal carcinoma patients treated with platinum-based chemoradiotherapy. Oncotarget 6, 28478–28490 (2015).
Sullivan, I. et al. Pharmacogenetics of the DNA repair pathways in advanced non-small cell lung cancer patients treated with platinum-based chemotherapy. Cancer letters 353, 160–166 (2014).
Arriagada, R. et al. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. The New England journal of medicine 350, 351–360 (2004).
Olaussen, K. A. et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. The New England journal of medicine 355, 983–991 (2006).
Du, H. et al. Association study between XPG Asp1104His polymorphism and colorectal cancer risk in a Chinese population. Scientific reports 4, 6700 (2014).
Zhou, B., Hu, X. M. & Wu, G. Y. Association between the XPG gene Asp1104His polymorphism and lung cancer risk. Genetics and molecular research: GMR 15 (2016).
Liu, D., Wu, J., Shi, G. Y., Zhou, H. F. & Yu, Y. Role of XRCC1 and ERCC5 polymorphisms on clinical outcomes in advanced non-small cell lung cancer. Genetics and molecular research: GMR 13, 3100–3107 (2014).
Therasse, P. et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. Journal of the National Cancer Institute 92, 205–216 (2000).
Friedberg, E. C. How nucleotide excision repair protects against cancer. Nature reviews. Cancer 1, 22–33 (2001).
Wu, W. et al. Effect of polymorphisms in XPD on clinical outcomes of platinum-based chemotherapy for Chinese non-small cell lung cancer patients. PloS one 7, e33200 (2012).
Wu, W. et al. Association of XPD polymorphisms with severe toxicity in non-small cell lung cancer patients in a Chinese population. Clinical cancer research: an official journal of the American Association for Cancer Research 15, 3889–3895 (2009).
Somers, J. et al. A common polymorphism in the 5′ UTR of ERCC5 creates an upstream ORF that confers resistance to platinum-based chemotherapy. Genes & development 29, 1891–1896 (2015).
Kim, S. H. et al. Clinical significance of ERCC2 haplotype-tagging single nucleotide polymorphisms in patients with unresectable non-small cell lung cancer treated with first-line platinum-based chemotherapy. Lung Cancer 77, 578–584 (2012).
Wu, X. et al. XPA polymorphism associated with reduced lung cancer risk and a modulating effect on nucleotide excision repair capacity. Carcinogenesis 24, 505–509 (2003).
Liu, X. et al. Association between XPA gene rs1800975 polymorphism and susceptibility to lung cancer: a meta-analysis. The clinical respiratory journal (2016).
Chen, P. et al. The functional status of DNA repair pathways determines the sensitization effect to cisplatin in non-small cell lung cancer cells. Cell Oncol (Dordr) 39, 511–522 (2016).
Dualan, R. et al. Chromosomal localization and cDNA cloning of the genes (DDB1 and DDB2) for the p127 and p48 subunits of a human damage-specific DNA binding protein. Genomics 29, 62–69 (1995).
Hu, Z. et al. Polymorphisms in DNA damage binding protein 2 (DDB2) and susceptibility of primary lung cancer in the Chinese: a case-control study. Carcinogenesis 27, 1475–1480 (2006).
He, L. et al. Two new susceptibility loci 1q24.2 and 11p11.2 confer risk to severe acne. Nature communications 5, 2870 (2014).
Barakat, B. M. et al. Overexpression of DDB2 enhances the sensitivity of human ovarian cancer cells to cisplatin by augmenting cellular apoptosis. International journal of cancer 127, 977–988 (2010).
Giglia-Mari, G. et al. Dynamic interaction of TTDA with TFIIH is stabilized by nucleotide excision repair in living cells. PLoS biology 4, e156 (2006).
Wang, M. et al. Genetic variant in DNA repair gene GTF2H4 is associated with lung cancer risk: a large-scale analysis of six published GWAS datasets in the TRICL consortium. Carcinogenesis 37, 888–896 (2016).
Buch, S. C. et al. Genetic variability in DNA repair and cell cycle control pathway genes and risk of smoking-related lung cancer. Molecular carcinogenesis 51(Suppl 1), E11–20 (2012).
Yi, Y. L. et al. XPG is a novel biomarker of clinical outcome in advanced non-small-cell lung cancer. Pak J Med Sci 29, 762–767 (2013).
Zou, H. Z. & Zhao, Y. Q. XPG polymorphisms are associated with prognosis of advanced non-small cell lung cancer treated with platinum-based doublet chemotherapy. Genetics and Molecular Research 14, 500–506 (2015).
Hu, W. C., Pan, J. B., Zhao, P., Yang, G. Y. & Yang, S. J. Genetic polymorphisms in XPG could predict clinical outcome of platinum-based chemotherapy for advanced non-small cell lung cancer. Tumor Biol 35, 5561–5567 (2014).
Zhang, T. et al. XPG is Predictive Gene of Clinical Outcome in Advanced Non-small-cell Lung Cancer with Platinum Drug Therapy. Asian Pac J Cancer P 14, 701–705 (2013).
He, C. Y., Duan, Z. P., Li, P., Xu, Q. & Yuan, Y. Role of ERCC5 promoter polymorphisms in response to platinum-based chemotherapy in patients with advanced non-small-cell lung cancer. Anti-Cancer Drug 24, 300–305 (2013).
Toledo, L. I. et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103 (2013).
Yuan, J., Ghosal, G. & Chen, J. The annealing helicase HARP protects stalled replication forks. Genes & development 23, 2394–2399 (2009).
Fang, C. et al. MiR-488 inhibits proliferation and cisplatin sensibility in non-small-cell lung cancer (NSCLC) cells by activating the eIF3a-mediated NER signaling pathway. Scientific reports 7, 40384 (2017).
Givalos, N. et al. Replication protein A is an independent prognostic indicator with potential therapeutic implications in colon cancer. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc 20, 159–166 (2007).
Wang, G. et al. PTEN regulates RPA1 and protects DNA replication forks. Cell research 25, 1189–1204 (2015).
Ma, H. et al. ERCC6/CSB gene polymorphisms and lung cancer risk. Cancer letters 273, 172–176 (2009).
Zhang, L. et al. Association between single nucleotide polymorphisms (SNPs) and toxicity of advanced non-small-cell lung cancer patients treated with chemotherapy. PloS one 7, e48350 (2012).
Nicolas, E., Golemis, E. A. & Arora, S. POLD1: Central mediator of DNA replication and repair, and implication in cancer and other pathologies. Gene 590, 128–141 (2016).
Valle, L. et al. New insights into POLE and POLD1 germline mutations in familial colorectal cancer and polyposis. Human molecular genetics 23, 3506–3512 (2014).
Dunlop, M. G. et al. Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nature genetics 44, 770–776 (2012).
Roe, O. D. et al. Molecular resistance fingerprint of pemetrexed and platinum in a long-term survivor of mesothelioma. PloS one 7, e40521 (2012).
Khor, C. C. et al. A Mal functional variant is associated with protection against invasive pneumococcal disease, bacteremia, malaria and tuberculosis. Nature genetics 39, 523–528 (2007).
Gochhait, S. et al. Implication of BRCA2 -26G > A 5′ untranslated region polymorphism in susceptibility to sporadic breast cancer and its modulation by p53 codon 72 Arg > Pro polymorphism. Breast cancer research: BCR 9, R71 (2007).
Acknowledgements
This research was funded by the Shanghai Municipal Commission of Health and Family Planning (20134Y193).
Author information
Authors and Affiliations
Contributions
Daru Lu, Baohui Han, Chunxue Bai, Qiang Li designed the study; Zhiqiang Gao, Ji Qian, Junjie Wu, Hongyan Chen collected the samples; Xiaoying Li, Xueying Zhao collected clinical information, Jiucun Wang, Cong Huai analyzed data; Xiao Song, Shiming Wang, Xuan Hong analyzed data and wrote the paper.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Song, X., Wang, S., Hong, X. et al. Single nucleotide polymorphisms of nucleotide excision repair pathway are significantly associated with outcomes of platinum-based chemotherapy in lung cancer. Sci Rep 7, 11785 (2017). https://doi.org/10.1038/s41598-017-08257-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-08257-7
This article is cited by
-
Immune characteristics analysis and construction of a four-gene prognostic signature for lung adenocarcinoma based on estrogen reactivity
BMC Cancer (2023)
-
A nuclease-mimetic platinum nanozyme induces concurrent DNA platination and oxidative cleavage to overcome cancer drug resistance
Nature Communications (2022)
-
CSB affected on the sensitivity of lung cancer cells to platinum-based drugs through the global decrease of let-7 and miR-29
BMC Cancer (2019)
-
Biological predictors of chemotherapy-induced peripheral neuropathy (CIPN): MASCC neurological complications working group overview
Supportive Care in Cancer (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.