Abstract
The lateral segregation of membrane constituents into functional microdomains, conceptually known as lipid raft, is a universal organization principle for cellular membranes in both prokaryotes and eukaryotes. The widespread Stomatin, Prohibitin, Flotillin, and HflK/C (SPFH) family proteins are enriched in functional membrane microdomains at various subcellular locations, and therefore were hypothesized to play a scaffolding role in microdomain formation. In addition, many SPFH proteins are also implicated in highly specific processes occurring on the membrane. However, none of these functions is understood at the molecular level. Here we report the structure of a supramolecular complex that is isolated from bacterial membrane microdomains and contains two SPFH proteins (HflK and HflC) and a membrane-anchored AAA+ protease FtsH. HflK and HflC form a circular 24-mer assembly, featuring a laterally segregated membrane microdomain (20 nm in diameter) bordered by transmembrane domains of HflK/C and a completely sealed periplasmic vault. Four FtsH hexamers are embedded inside this microdomain through interactions with the inner surface of the vault. These observations provide a mechanistic explanation for the role of HflK/C and their mitochondrial homologs prohibitins in regulating membrane-bound AAA+ proteases, and suggest a general model for the organization and functionalization of membrane microdomains by SPFH proteins.
Similar content being viewed by others
Introduction
Cellular membranes are extremely heterogeneous in composition, with a non-random distribution of lipids and proteins. One universal feature of cellular membranes from all three kingdoms of life is the presence of laterally segregated nanoscale microdomains, serving as compartmentalized platforms for many essential functions of the cells. The organization and regulation of these functional membrane microdomains (FMMs) are best studied for the eukaryotic plasma membrane, and “lipid raft”,1 as a well-accepted model, is to describe these relatively ordered discrete microstructures formed through interactions among enriched cholesterol, sphingolipid and a certain type of proteins.1,2,3 Although bacteria generally do not contain sterol lipids, their membranes are similarly organized into microdomains4,5 that also require FMM-specific lipid species.6 As maker proteins, SPFH (Stomatin, Prohibitin, Flotillin, and HflK/C) family proteins are enriched in prokaryotic and eukaryotic FMMs of various subcellular localizations, including plasma membrane, nucleus, Golgi apparatus, endoplasmic reticulum (ER), endosomes, lipid droplets, and mitochondria (reviewed in7,8,9,10). The common property of SPFH proteins in self-oligomerizing into large membrane-spanning or membrane-anchored complexes suggests a potential scaffolding role for them in the formation and organization of FMMs.7,11
Apart from this general function, owing to their ability to interact with distinct protein partners, SPFH family members possess very specific regulatory roles in highly diverse cellular processes. For example, mammalian flotillins are present in plasma membrane, endosomes and exosomes, and regulate various signal transduction and membrane trafficking events.12,13,14 Similarly, bacterial flotillins are also abundant and colocalize with a wide range of cargo proteins in FMMs,15 including those involved in cell wall synthesis required for penicillin resistance in methicillin-resistant Staphylococcus aureus (MRSA).16 A few members of SPFH family proteins display highly specialized functions: C. elegans stomatin MEC-2 and its mouse homolog stomatin-like protein 3 (SLP3) are implicated in mechanosensation by linking acid-sensing ion channels to cytoskeleton,17,18,19 whereas human erlins20 interacts with components of ER-associated degradation (ERAD) pathway and a ubiquitin ligase RNF170 to mediate the degradation of inositol trisphosphate receptor (IP3R).21,22
Unlike these cellular SPFH proteins, prohibitins (PHB1/2) predominantly exist in the inner membrane of mitochondria.23,24 PHB1 was first identified as a potential tumor suppressor,25 and since then prohibitins have been demonstrated to be important for various aspects of mitochondrial dynamics and metabolisms, as well as for cell survival and proliferation (reviewed in26,27). Prohibitin-targeting small molecules also displayed promising effects against cancer and neurodegenerative, metabolic, and inflammatory diseases.28 PHB1/2 and their bacterial homologs HflK/C share a common function in regulating membrane protein quality control and homeostasis through direct interactions with hexameric AAA+ membrane-bound proteases Yta10/Yta1224 and FtsH,29 respectively. Therefore, many of the pleiotropic effects of prohibitins on physiology and pathology could be explained by the diverse substrates of the mitochondrial AAA+ protease.30,31,32,33,34
However, none of the roles of SPFH family proteins, either in general FMM scaffolding or in specialized regulatory functions, is understood at molecular level. Previous structural studies on fragments of different SPFH members, including SPFH domains of mouse stomatin,35 Pyrococcus horikoshii stomatin,36,37 and Vibrio alginolyticus FliL,38 as well as a coiled-coil region of human PHB2,39 provided sharply different inter-subunit arrangements, leading to conflicting assembly models for high-order oligomerization. Importantly, low-resolution electron microscopy data revealed a ring-shaped hollow assembly for PHB1/2 complex from Saccharomyces cerevisiae40 and also for a cyanobacterial stomatin homolog.41 These early observations allowed the formulation of an attractive model that prohibitins self-oligomerizes to form a sequestered space for certain biological processes.40 Consistently, mammalian major vault protein (distantly related to SPFH family), which is a cytoplasmic protein but recruited to lipid rafts after Pseudomonas aeruginosa infection,42 also displays a circular assembly comprising 39 laterally arranged subunits.43 The copy number of PHB1/2 heterodimer in the prohibitin complex was estimated to be 12 to 16,44 whereas a number of 6 to 10 was proposed for HflK/C.45,46 Therefore, the exact composition and subunit arrangement of membrane-spanning SPFH complexes, as well as their potential interactions with other proteins, remain largely unclear.
In the present work, E. coli HflK and HflC were employed as a model system for structural characterization of SPFH family proteins. Similar to mitochondrial prohibitins, HflK and HflC copurify with membrane-bound FtsH complex and regulate its proteolytic activity on both cytoplasmic and membrane substrates.29,46,47,48 Thus, we prepared a complex formed by HflK, HflC and FtsH, and determined a high-resolution structure of this membrane supra-assembly using cryo-electron microscopy (cryo-EM). The 2.7 MDa structure display a gigantic architecture, bordered by 12 copies of HflK–HflC dimer to provide a large, compartmentalized periplasmic cage and a laterally segregated, but cytoplasm-exposed membrane area with four embedded FtsH hexamers. These features suggest a straightforward model for the FMM organization that SPFH proteins oligomerize into nanometer-sized circular assemblies to facilitate the compartmentation of lipids and membrane proteins.
Results
HflK–HflC–FtsH complex preparation and structural determination
Overexpressed HflK, HflC and FtsH proteins were purified from solubilized membrane fractions of E. coli BL21 (DE3) cells through a FLAG tag at the C-terminus of HflC. Different detergents and varying concentrations of glycerol (from 0% to 10%) in purification buffers were tested and evaluated by negative-staining electron microscopy (nsEM) (Supplementary information, Fig. S1a–h), and a combination of n-Dodecyl-β-d-Maltoside (DDM) and 8% glycerol was finally used for complex preparation. To further increase the sample homogeneity, a glycerol density gradient centrifugation was applied (Supplementary information, Fig. S1j) and peak fractions were used for functional and structural characterization. The distribution of HflK and HflC in the glycerol fractions is very wide, and each fraction was examined using nsEM (Supplementary information, Fig. S1k, l). Intact HflK–HflC–FtsH (KCF) complexes were subsequently subjected to a protein degradation assay using two reported substrates of FtsH, a soluble bacteriophage λ CII protein47 and a membrane protein SecY.29 Purified KCF complex is competent in degrading both substrates, and as expected, it is less active on detergent-solubilized SecY (Supplementary information, Fig. S2).
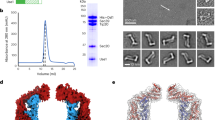
The nsEM analysis revealed very interesting features for the KCF complex, resembling a circular cage-like structure (Supplementary information, Fig. S1m). Subsequent cryo-EM analysis confirmed this observation and revealed that KCF particles are highly heterogenous in both composition and conformation (Fig. 1). A source of heterogeneity was likely from sample purification and grid preparation. Many classes from 3D classification show large ruptures in the structures and are deformed to different extents (Supplementary information, Figs. S3, S4). In addition, FtsH hexamers are present in different copy numbers in the KCF complexes, ranging from zero to four (Fig. 1; Supplementary information, Fig. S4). The inclusion of FtsH hexamers leads to a shape transition of the HflK/C enclosure from circular to square-like (Fig. 1a–d). Therefore, for high-resolution refinement, only those intact particles (less than 10% of all particles) with four FtsH hexamers from multiple datasets were combined for refinement with C4 symmetry imposed (Supplementary information, Fig. S3e), which led to a final density map for the overall complex at 3.3 Å resolution (Gold-standard Fourier Shell Correlation 0.143 cutoff) (Supplementary information, Fig. S5d, e). Atomic models could be built de novo in this global map for most of the sequences of HflK and HflC (Supplementary information, Fig. S5f–h), except their transmembrane (TM) helices and the regions proximal to the membrane. The density of these regions was further improved by focused classification and refinement, which allowed subsequent atomic modeling of these regions, including the upper portion of TM helices (Supplementary information, Figs. S3e and S5i, j).
The overall structure of the KCF complex resembles an inverted cup on the membrane, with the cup wall formed by circularly arranged 12 copies of HflK–HflC dimers (Fig. 2). Consistent with the previously determined topology of these proteins,47,49 most sequences of HflK/C locate in the periplasm, whereas the two-layered catalytic domains of FtsH are anchored on the cytoplasmic side of the membrane (Fig. 1). In the high-resolution maps, the density of cytoplasmic domains of FtsH becomes highly fragmented, but its hexameric periplasmic region remains stable (Fig. 2b) and matches well with the crystal structure of this region (residues 30–92) (PDB 4V0B).50 From the side view, the KCF structure could be divided into three parts, the TM, shoulder, and cap regions (Fig. 2c). The periplasmic domains of the four FtsH hexamers are localized completely inside the cup, with specific interactions with the inner surface of the HflK/C wall in the shoulder region. The inclusion of four FtsH hexamers within the HflK/C wall results in a symmetry mismatch between the cap (C12) and the shoulder (C4) regions.
a Top view (periplasmic view) of the cryo-EM map (left) and atomic model (right) of the KCF complex. b Bottom view (cytoplasmic view) of the cryo-EM map (left) and atomic model (right) of the KCF complex. c Side view of the cryo-EM map (left) and atomic model (right) of the KCF complex. Subunits of HflK, HflC and FtsH are colored cyan, magenta and orange, respectively.
Structures of HflK and HflC and their transmembrane domains
Both HflK and HflC are typical single-pass type II membrane proteins, with a single N-terminal TM helix. Although the sequences of HflK and HflC are largely diverged (only 13% sequence identity), they share a highly conserved 3D structure. Following the TM helix (α1), both structures could be divided into four domains, two conserved SPFH domains (SPFH1 and SPFH2), a coiled-coil domain (CC1 and CC2), a C-terminal domain (CTD) (Fig. 3a, b).
a, b Schematic illustration of the conserved domain organization of HflK and HflC, with individual domains separately colored. TM, blue; SPFH1, cyan; SPFH2, green; CC1, yellow; CC2, orange; CTD, red. The N-terminal and C-terminal extensions of HflK are denoted by NTE and CTE, respectively. c, d The atomic model (c) and the secondary structure topology of HflK (d). Secondary structural motifs are colored as in a. e, f The atomic model (e) and the secondary structure topology of HflC (f). Secondary structural motifs are colored as in b. g SPFH1 domains of HflK and HflC are peripheral membrane domains inserted in the outer leaflet of the membrane. The low-pass filtered cryo-EM map of the KCF complex is displayed in transparent surface representation, with atomic models of HflK (cyan) and HflC (magenta) superimposed. Detergent density is colored dodger blue. h Zoomed-in view of the TM regions of the KCF complex. i Same as h, with the removal of detergent density in the foreground. Two aromatic residues (F34 and F58 for HflC; F111 and F128 for HflK) located in the β2–β3 and β4–β5 loops are inserted in the membrane. j, k Vertical cross-section view of the density map of the KCF complex, in the position of the TM (j) and SPFH1 (k) regions. l, m The distribution of charged residues in the SPFH1 domains of HflK (l) and HflC (m). Note that membrane-buried polar residues often appear in pairs of opposite charge. R33 of HflC and R110 of HflK are highly conserved among SPFH family proteins from both prokaryotes and eukaryotes (see also Supplementary information, Fig. S6a).
The structures of SPFH1 domains have not been previously reported for membrane-associated SPFH proteins, except for the soluble MVP from rat which contains 9 tandemly arranged SPFH1 domains.43 An important finding is that the SPFH1 domains of HflK and HflC, composed of five antiparallel β-strands (β1–β5) (Fig. 3c–f), are in fact peripheral membrane domains inserted in the outer leaflet of the membrane, with nearly 1/2 of the mass buried in the detergent micelle (Fig. 3g–i). The SPFH1 domains contain many hydrophobic residues, including 5 and 6 aromatic residues for HflC and HflK, respectively (Supplementary information, Fig. S6a). Some of them apparently play an important role in membrane insertion. Two examples are the phenylalanine-containing loops located between β2 and β3, and between β4 and β5. These two loops are inserted into the outer leaflet in different depths, and their phenylalanine side-chains are both perpendicular to the membrane plane (Fig. 3i). Furthermore, a few highly conserved charged residues of SPFH1 domains are exactly next to the membrane surface (Fig. 3l, m; Supplementary information, Fig. S6a). Of note, the membrane plane in the detergent micelle appears to have a curvature (Fig. 3g), suggesting a possibility of the HflK/C complex in membrane reshaping. Very importantly, the 24 SPFH1 domains together with N-terminal TM helices define a membrane area of 300–400 nm2 that is isolated from surroundings (Fig. 3j, k). It must be noted that many disease-related mutations of human podocin, a SPFH member implicated in hereditary steroid-resistant nephrotic syndrome,51,52 were found in the SPFH1 domain (Supplementary information, Fig. S6a). This highlights the physiological significance of the SPFH1 domain in membrane compartmentation.
The structures of SPFH2 domains are very similar to previously reported fragment structures of other SPFH family members,35,36,37,38,43 with an α/β-fold of three β-strands and four α-helices. The following coiled-coil domains of HflK and HflC are long α-helices (94 residues), which are divided into two pieces, CC1 (α6) and CC2 (α7), by a 45°-bending in the C-terminal one third position (residues of P291 and P319 for HflC and HflK, respectively). Right-handed helical packing of CC1 domains from HflK and HflC leads to the formation of the cylinder wall of the KCF complex, whereas CC2 domains and the CTDs form the flat top of the cap (Fig. 3c–f).
HflK and HflC differs from each other in several insertion sequences. Firstly, HflK possesses an additional N-terminal extension (NTE, residues 1–78), which contains 23 glycine residues. This cytoplasmic extension is highly flexible and not resolved in the map. Secondly, HflC contains two insertions in its SPFH1 and SPFH2 domains. One is a short β-hairpin between the β2 and β3 of the SPFH1 domain (Fig. 3e, f; Supplementary information, Fig. S6a), and the other is an insertion (residues 165–201, not resolved in the density map) in the α5 of the SPFH2 domain (Fig. 3e, f; Supplementary information, Fig. S6b). Thirdly, the sequences and structures of the CTDs of HflK and HflC are sharply different (Fig. 3c–f). The CTD of HflC starts with a short β-strand (β9), followed by a helix (α8), and the C-terminus is exposed in the periplasmic space. In contrast, the CTD of HflK is characterized by an additional β-strand (β10) following β9, which turns around and places the C-terminal extension (CTE, residues 351–419) of HflK inside the KCF complex. This CTE is also highly flexible and not fully resolved in the map.
Oligomerization of HflK and HflC
The 12 copies of HflK and HflC (numbered as C1–C12 and K1–K12) perfectly align from the N-terminus to the C-terminus to form a tightly sealed cage in the periplasmic space (Figs. 2, 4). Owing to the interaction with four FtsH hexamers (Fig. 4a), they form 6 unique heterodimeric interfaces (Supplementary information, Fig. S7a-f). Three C–K interfaces (C1–K1, C2–K2 and C3–K3) are apparently tighter, whereas two of the three K–C interfaces (K2–C3, K3–C4) display an apparently larger distance between adjacent SPFH domains (Fig. 4a, b). Comparison of the three C–K interfaces using the CC2 subdomains as reference of alignment shows that the SPFH1 and SPFH2 domains display displacements at varying extents with root-mean-square deviation (RMSD) ranging from 3.0 to 9.3 Å (Supplementary information, Fig. S7g–i). In contrast, if the SPFH1 and SPFH2 domains of HflC were used as reference for the interface alignment, SPFH1 and SPFH2 domains of HflK could be well superimposed (RMSD 1.0–1.4 Å) (Supplementary information, Fig. S7j–l), indicating that C–K interfaces are relatively rigid in the region of SPFH domains. As for the three K–C interfaces (K1–C2, K2–C3 and K3–C4), similar analyses reveal much larger structural differences (Supplementary information, Fig. S7m–r), and the contact surfaces between adjacent HflK and HflC SPFH domains are also different (Supplementary information, Fig. S7p–r). These results imply a larger conformational plasticity for the K–C interfaces, and also suggest that the relatively stable C–K dimer likely represents a basic assembly unit of HflK–HflC complex.
a A cutaway view of the KCF complex (viewed from the periplasmic side). HflK, HflC and FtsH are colored cyan, magenta and orange, respectively. Protomers of HflK (K1 to K12) and HflC (C1 to C12) are numbered anti-clockwisely. b, c Side view of a quarter of the KCF complex (one unsymmetrical unit). Subunits of HflK, HflC and FtsH are similarly colored as in a. d Domain interface between C1 and K1 in the SPFH1 region. The cryo-EM maps of the SPFH1 and SPFH2 domains are shown in transparent surface representation with atomic models superimposed. In the C1–K1 interface, the F111–G112–K113 loop (β2–β3 loop) of HflK contacts two loops (β1–β2 and β3–β4 loops) of HflC–SPHF1. e Detailed interactions between adjacent SPFH1 domains in the C1–K2 interface, including a hydrogen bond between main-chain atoms of G112 of K1 and K24 of C1, and a hydrophobic stacking between L52 of C1 and F111 of K1. f Same as d, but for the K1–C2 interface. In the C1–K2 interface, the F34–G35–K36 loop (β2–β3 loop) of HflC contacts two loops (β1–β2 and β3–β4 loops) of HflK-SPHF1. g Detailed interactions between adjacent SPFH1 domains in the C1–K2 interface, including a hydrogen bond between main-chain atoms of G35 of C2 and K101 of K1, and a hydrophobic stacking between L122 of K1 and F34 of C2. Note that the three interacting loops share identical sequences between HflK and HflC (See also Supplementary information, Fig. S6a). h Electrostatic surface potential of the interface between the SPFH2 domains of HflC (C1) and HflK (K1). i The C1–K1 interface is shown in cartoon representation, with interacting residues highlighted in stick models. j–o Same as h and i, for the K1–C2 (j, k), K2–C3 (l, m) and K3–C4 (n, o) interfaces.
The tight association between HflK and HflC results from a combination of interfaces on their juxtaposed domains. Taking the interfaces of C1–K1 and K1–C2 as examples, a general principle governing the assembly of HflK and HflC could be deduced. The interactions between adjacent SPFH1 domains are at the membrane interface and mediated by inter-strand loops on their β-strand backbones (Fig. 4d–g). In the C1–K1 interface, a glycine-containing loop of HflK (F111–G112–K113, β2–β3 loop) contacts two loops of HflC-SPHF1 (β1–β2 and β3–β4 loops), capable of forming hydrogen bonds through their main-chain atoms (Fig. 4e). In addition, a hydrophobic stacking between HflC-L52 and HflK-F111, both of which are in fact inserted in the outer leaflet of the membrane, also contributes to this interface. This pattern of interactions is highly similar in the K1–C2 interface: The equivalent β2–β3 loop (F34–G35–K36) of HflC exactly interacts with the two equivalent loops of HflK (Fig. 4g). Notably, HflK and HflC share identical sequences in these three interacting loops (Supplementary information, Fig. S6a). Therefore, the SPFH1 interface offers no distinction in selecting HflC or HflK for oligomerization. As to the SPFH2 domains, the interface is apparently mediated by oppositely charged surface patches between HflC and HlfK (Fig. 4h–o). A clear charge complementarity could be found in the three relatively rigid C–K interfaces (Fig. 4h, i). The same holds true for the three K–C interfaces, although different surface areas are used (Fig. 4j–o). Importantly, the constellation of the charged patches on the two sides of HflC and HflK determines that the two molecules must bind in alternative order to assemble a complete complex.
The extensive atomic contacts between paralleled CC1 subdomains are distributed over the entire helices and show no bias toward polar (Supplementary information, Fig. S8a–h) or hydrophobic interactions (Supplementary information, Fig. S8l–r), but with necessary side-chain reconfiguration in different C–K and K–C interfaces (Supplementary information, Fig. S8i–k). The formation of the cap of the KCF complex, in contrast, is predominately mediated by hydrophobic interactions (Fig. 5). For example, the C-terminal helix (α8) of HflC is amphipathic, which is exactly docked on the juxtaposed CC2 helices using its hydrophobic surface (Fig. 5b, c). Following CC2 helices, the C-terminal sequences of HflC and HflK constitute a two-layered β-barrel, a 24-stranded (β9 of HflK and HflC) outer layer and a 12-stranded inner layer (β10 of HflK) (Fig. 5d–g). Notably, this arrangement leaves a central channel (15 Å in diameter) in the cap, allowing the access to the KCF chamber from the periplasmic space.
a Top view of the KCF complex, with seven subunits highlighted in ribbon representation and separately colored (C1, K1, C2, K2, C3, K3 and C4). b, c Zoomed-in view of boxed region in a. Extensive hydrophobic interactions contribute to the formation of the cap region (b). Detailed hydrophobic interactions between the α8-helix of HflC and juxtaposed CC2 subdomains of HflK and HflC (c). d Top view of the KCF complex, highlighting the overall organization of the cap region. e Enlarged view highlighting the positions of the C-termini of HflK and HflC. f, g Top (f) and side (g) views of the two-layered β-barrel in the cap region of the KCF complex. The outer layer is formed by 24 parallel β9-strands from HflK and HflC, and the inner layer by 12 parallel β10-strands of HflK.
Interactions between FtsH and HflK
Although FtsH stably associates with HflC and HflK during purification, owing to the glycine-rich linker between its transmembrane region and the AAA+ module, only periplasmic domains (PDs) of FtsH are well resolved in the density map. Moreover, it displays unexpectedly limited interactions only with HflK. One FtsH hexamer is observed to interact with multiple copies of HflK (Fig. 6a–f). Three PDs of a single FtsH hexamer are arranged in a similar orientation towards three neighboring HflK subunits, and two of them establish highly specific interactions, through a β-hairpin loop (K61–D62–S63–N64, β2–β3) with the linker sequence (R141–E142) between SPFH1 and SPFH2 domains of HflK (K1 and K12). The two atomic interfaces are generally similar, involving a few highly charged residues from two parties and two polar residues (R185 and D181) from the α3-helix of SPFH2 (Fig. 6c, f). Notably, although the atomic arrangements of the two interfaces are different in details, they share a common set of main-chain interactions: the carbonyl oxygens of K61 and D62 form hydrogen bonds with the main-chain nitrogen of E142 and the side-chain of R141, respectively; S63 also interacts with R141 through their side-chains. To verify this structurally resolved interface, we introduced alanine mutations at selected positions and examined their impacts on the binding of FtsH (Fig. 6j). Unexpectedly, most single interface mutations, including HflK-R141A, FtsH-D62A, FtsH-N64A, FtsH-S63A and FtsH-K61A had no or merely moderate effect on the stability of the KCF complex, except HflK-E142A, which alone resulted in significantly reduced binding of FtsH. This suggests that the strength of the observed interface is determined by a combination of polar interactions. Indeed, simultaneous mutations of multiple interface residues, led to dramatically impaired FtsH association (Fig. 6j).
a Interaction between an FtsH hexamer and the inner wall of the HflK/C cage within a quarter of the KCF complex. Three FtsH periplasmic domains (F1, F2 and F6) are similarly orientated towards three copies of HflK (K1, K12 and K2), and two of them establish specific interactions. b, c Zoomed-in view showing the interactions between F1 and K1. The β2–β3 loop of FtsH contacts the linker sequence between the SPFH1 and SPFH2 domains of HflK (b). Detailed atomic interactions are highlighted in stick representation with distances labeled (c). d Same as a, with a further zoom in the two interacting FtsH periplasmic domains. e Structural comparison of the two FtsH–HflK interfaces (F1–K1 and F2–K12). Structural superimposition of F1–K1 and F2–K12 is derived using FtsH periplasmic domains as reference for alignment. F1 and K1 are colored gray. F2 and K12 were colored orange and cyan, respectively. f Same as c, but for the interface between F2 and K12. g Top view of the cryo-EM map of the KCF complex, highlighting the unmodelled density (cyan) above the hexameric periplasmic domain of FtsH. h Same as g, with detergent density removed. i Segmented cryo-EM map of a hexameric periplasmic domain of FtsH. j Point mutations on FtsH or HflK impair the binding of FtsH to the HflK/C cage. For each pull-down experiment with mutant variants, the C-terminally tagged HflC was used as bait and the loading was normalized using concentrations of HflC. k Truncations of HflK C-terminal sequences decrease the association of FtsH. For each pull-down experiment with mutant variants, the C-terminally tagged HflC was used as bait and the loading was normalized using concentrations of HflC.
Nevertheless, the disruption of the interface could not completely abolish the binding of FtsH to the HflK/C cage, which suggests the presence of additional interactions that are not fully resolved in high-resolution maps. A careful examination of the map revealed a ring-shaped density above the hexamer of FtsH PDs (Fig. 6g–i). Since the CTE of HflK ends in the cage, we constructed a set of progressive truncations from the C-terminus of HlfK and tested their effects on FtsH binding (Fig. 6k). The deletion of the last 10 residues (Δ410–419) alone is sufficient to reduce the co-purified FtsH to a very low level. Further trimming of the C-terminal residues until the end of β9-strand (Δ347–419) almost completely abolished the binding of FtsH. In sharp contrast, removal of the N-terminal glycine-rich extension of HflK (Δ1–70), or the SPFH2 insertion of HflC (Δ160–190) had no effect in FtsH association (Fig. 6k).
These results indicate that the assembly of FtsH into the KCF complex is mediated by interactions with HflK. Considering the varying stoichiometry of FtsH hexamers in different KCF complexes (Fig. 1), the flexible CTE of HflK could play a major role in recruiting FtsH into the assembling cage of HflK/C.
Discussion
Membrane protein quality control by the KCF complex
FtsH is conserved in bacteria, mitochondria and chloroplasts, and plays a role in many cellular pathways through regulated proteolysis of certain proteins.53 One function of bacterial FtsH is, as a component of stress response circuits, to degrade cytosolic proteins including the heat-shock transcription factor σ32,54 a key enzyme LpxC in lipid A biosynthesis,55 and a phage transcription factor λCII56 which regulates lysis/lysogeny decision of phage λ. Its mitochondrial homologs form two types of FtsH-related AAA-proteases (m-AAA and i-AAA proteases), both of which are anchored to the mitochondrial inner membrane. While the m-AAA protease has a similar topology as bacterial FtsH, the i-AAA protease exposes its enzymatic domains in the intermembrane space.57 Mutations in human FtsH homologs AFG3L2 and SPG7 are associated with neurological disorders.31,58,59,60 The functional diversity of FtsH and its mitochondrial counterparts are largely due to their ability to process different membrane protein substrates. For examples, bacterial FtsH proteases share a few common substrates, such as SecY of the SecYEG translocase and F0a subunit of the ATP synthase, as well as a few species-specific membrane protein substrates (reviewed in53).
The architecture of the KCF holoenzyme intermediately suggests a straightforward model that HflK and HflC limit the substrate access to FtsH hexamers by confining the enzyme in a laterally segregated space (Fig. 7a, b). The HflK/C complex therefore represents an additional layer of regulation on the proteolytic activity of FtsH. This model is consistent with the view of HflK/C as a modulator of FtsH.48 The HflK/C complex negatively regulates the protease activity of FtsH against membrane proteins, such as SecY and YccA, as null mutation of HflK/C accelerated the degradation of SecY in vivo.29 In support of our structural findings, deletion of the periplasmic region of FtsH, which would disrupt the interaction between FtsH and HflK, did influence the in vivo proteolytic activity of FtsH in degrading these membrane substrates.61 Similar to HflK/C, loss of PHB1 or PHB2 in yeast also accelerated proteolysis of non-assembled mitochondrial inner membrane proteins by m-AAA proteases.24
a FtsH hexamers degrade misfolded/damaged membrane proteins or cytoplasmic proteins. b HflK and HflC form a 24-mer circular assembly on the membrane, leading to the formation of 20-nm-sized microdomains bordered by the N-terminal TM domains of HflK/C. Up to four FtsH hexamers could be sequestered in these laterally segregated microdomains. HflK and HflC function as a negative modulator of FtsH to limit its substrate accessibility. c A general model for the FMM formation by SPFH family proteins. HflK/C, erlins and prohibitins contain a N-terminal TM segment, whereas stomatin, podocin and eukaryotic flotilins attach to the inner leaflet of the membrane through a hairpin helix or solely through lipidation of selected N-terminal residues. In all these cases, oligomerization of SPFH domains result in the formation of nanoscale microdomains on specific leaflets of the membranes.
Over one fifth of bacterial genes encode membrane proteins, creating a rather crowded surrounding for FtsH proteases. Therefore, it is necessary for the cells to limit the access of FtsH to functional membrane proteins, which underlines the physiological importance of HflK/C in vivo. Apparently, the presence of a large periplasmic HflK/C cage would prevent membrane proteins or complexes, especially those with large periplasmic domains, from getting close to FtsH. In addition, the tight interactions between the adjacent membrane-inserting SPFH1 domains of HflK/C result in a complete isolation of an area of the outer leaflet, which would stop the diffusion of any TM-containing membrane proteins into the cage, including those with a single TM helix. Furthermore, the 24 TM helices of the HflK/C complex could also act as a sieve to limit the diffusion of those peripheral or lipid-anchored proteins on the inner leaflet of the membrane. Therefore, one evident role of the HflK/C complex is to safeguard the membrane proteome from undesired proteolysis.
More importantly, the architecture of the KCF complex also explains the mechanism of FtsH’s role in membrane protein quality control. To be more specific, the isolation of the FtsH protease by an HflK/C enclosure provides a means to distinguish defective/faulty membrane proteins from functional ones. Damaged or uncomplexed membrane proteins are more likely to have flexible unfolded/misfolded termini which could be selectively recognized by the KCF holoenzyme. This agrees with that fact that FtsH preferentially degrades SecA and F0a when they are not in the context of functional complexes.62,63 Although hflK and hflC are not essential genes, when cells are challenged by stress conditions that lead to protein misfolding, the function of HflK/C in regulating FtsH would become more important. For example, ΔhflK and ΔhflC cells are more sensitive to aminoglycoside antibiotics, which induce mistranslation and consequently misfolding/mis-assembly of membrane proteins.64 In fact, the genes encoding HflK and HflC, as well as FtsH, are all under control of heat-shock promoters.65 Their joint involvement in the heat-shock response suggests that the role of FtsH in membrane protein quality control ought to be fulfilled in the form of the KCF holoenzyme. In addition, the varying stoichiometry of FtsH hexamers in the KCF complexes (Fig. 1) further suggest that the relative cellular levels of HflK/C and FtsH is likely controlled through various regulatory circuits, reflecting the cellular needs to maintain the homeostasis of membrane proteins.
Functional membrane microdomain organization by SPFH proteins
Over 67,000 SPFH domain-containing sequences across kingdoms have been identified according to the SMART database.66 However, these sequences display relatively low similarities among subgroups,67,68 especially for those eukaryotic members. This unclear phylogenetic relationship has limited functional studies in revealing precise functions of SPFH proteins. Our structure of the full-length KCF complex represents the first example of membrane-associated SPFH protein, which could serve as a structural reference for all SPFH family members. Therefore, guided by secondary structural prediction, we could now produce a unified model for the domain organization of diverse SPFH proteins from different subcellular locations (Supplementary information, Fig. S9). The three core features of SPFH proteins include two structurally conserved domains, SPFH1 and SPFH2, and a long α-helix (CC domain). While the two SPFH domains are in the range of 40–50 and 100–120 amino acids on average, the CC domains vary largely in length and can be divided into two subdomains (CC1 and CC2). Immediately after CC2, a short β-strand lies in the C-terminal sequences of all SPFH proteins (Supplementary information, Fig. S6c), which is equivalent to the β9 strands of HflK/C (Fig. 3). This suggests that other SPFH complexes also contain a similar β-barrel formed by these β-strands. The secondary structural motifs from SPFH1 to the C-terminal β-strand (the β9 equivalent of HflK/C) are the signature for all SPFH family proteins. The two termini are the most divergent regions. The C-termini after the β9-strand vary in size and could be either exposed (HflC-like) or buried inside the cage (HflK-like). The N-termini usually contain a hydrophobic helix that serves as a TM segment or membrane insertion hairpin. Crucially, our data in fact indicate that the SPFH1 domain of SPFH family proteins together with the N-terminal hydrophobic helix constitutes the membrane domain. An exception is eukaryotic flotillins, which lack the N-terminal hydrophobic helix and associate with the membrane solely through lipidation of selected N-terminal residues.7,11,69,70 Of note, some N-terminal TM helix-containing SPFH members display completely opposite topology from HflK/C, with the cage exposed in the equivalent compartment of cytosol.71,72 Nevertheless, the high conservation in their domain organization indicates that all SPFH proteins should share a common mechanism of oligomerization by an end-to-end juxtaposition of a certain number of subunits to form a circular structure on the membrane (Fig. 7c).
Together with the literatures on the biochemical and functional properties of SPFH proteins, our structural data allow the formulation of a straightforward model for understanding the primary function of SPFH proteins, as well as the roles of their individual domains, in organizing functional membrane microdomains. Firstly, SPFH proteins oligomerize to form a large assembly on the membrane, resulting in laterally compartmented membrane domains that could serve as platforms for specific membrane-related processes. Distinct SPFH proteins of various membrane fractions, presumably with different subunit stoichiometry in homo- or hetero-oligomerization, can organize FMMs in different physical sizes. Interestingly, the HflK/C complex sequesters a circular membrane area of 20 nm in diameter, which is compatible with the reported size of lipid rafts in eukaryotes (10–200 nm and 26 nm on average in diameter).3,73,74 This suggests that SPFH proteins likely define the physical boundary of certain types of FMMs.
Secondly, SPFH proteins may have an active role in binding and recruiting FMM-specific lipid species. Although the properties of specific lipid interaction for most SPFH proteins have not been characterized, a few members, including Caenorhabditis elegans MEC-2, mouse podocin and human stomatin were found to bind cholesterol,75,76 and a potential cholesterol-recognition/interaction amino acid consensus (CRAC) motif in the SPFH1 domain of human stomatin has been characterized.75 Cholesterol is one of the major lipid species in eukaryotic rafts.1,3,77 Mitochondrial membranes have a high level of cardiolipin and phosphatidylethanolamines and yeast PHB1/2 complex was proposed to promote the formation of membrane domains that are enriched in these two lipids.78 Bacterial membranes also do not contain cholesterol; instead, Staphylococcus aureus flotillin interacts with isoprenoid lipids, such as staphyloxanthin, in detergent-resistant membrane (DRM) fractions.16 Importantly, by forming an isolated membrane microdomain, SPFH proteins could also limit the lipid exchange across the microdomain. In the case of HflK/C, the diffusion of lipids on the outer leaflet into the microdomain is prohibited, whereas the diffusion within the inner leaflet is possible. Therefore, through inserting their SPFH1 domains into different leaflets of the membrane, SPFH proteins with different topology could regulate the local fluidity of distinct leaflets (Fig. 7c).
Thirdly, SPFH proteins directly interact with a variety of FMM cargo proteins, and may have a broad role in functionalizing membrane microdomains, with some of them displaying highly specialized regulatory functions in certain membrane-related processes. In addition to the well characterized examples of HflK/C, prohibitins and erlin1/2 that regulate membrane protein degradation, several SPFH members, including stomatin, podocin, SLP-3 and MEC-2, were reported to bind ion channels and regulate their activities.17,19,76,79 For another example, flotillin from gram-positive bacteria specifically interacts with PBP2a, an enzyme involved in cell wall synthesis.16 Our structure of the KCF complex suggests that other SPFH proteins may form similar super membrane complexes with these partners to physically isolate them to modulate their functional activities.
In summary, the structure of the KCF holo-enzyme explains the molecular role of HflK/C in regulating proteolysis of membrane proteins, and provides a framework for understanding pleiotropic functions of disease-relevant prohibitins and mitochondrial m-AAA proteases. More importantly, our study reveals the structural basis of FMM formation by SPFH proteins, and allows the formulation of interesting and testable models of different SPFH members for their general function in lipid and membrane protein compartmentation, as well as for their specific roles in regulating activities of various FMM cargoes.
Materials and methods
Cloning, expression and purification of the HflK–HflC–FtsH complex
E. coli strains were cultured in LB broth at 37 °C. E. coli DH5α was used for DNA manipulation, and E. coli BL21 (DE3) and E. coli C43 (DE3) were used for protein overexpression and purification. Antibiotics were used as following: ampicillin, 100 μg/mL; kanamycin, 25 μg/mL. For KCF and SecY expression, 0.2 mM IPTG was added to induce protein expression overnight at 18 °C. For CII expression, 0.5 mM IPTG was added to induce protein expression at 37 °C for 4 h.
Three genes of hflK, hflC and ftsH were amplified individually from E. coli DH5α genomic DNA by PCR. Genes of hflK and hflC were subsequently cloned into pETDuet and ftsH into pRSFDuet plasmids. For purification purpose, a Strep tag was inserted at the N-terminus of HflK; an 8× His tag and a Flag tag were attached to the C-termini of FtsH and HflC, respectively. Two recombinant plasmids (pETDuet-hflK-hflC and pRSFDuet-ftsH) were transformed into E. coli BL21 (DE3) cells for expression. When cells grew to a density of 1.0 at 600 nm at 37 °C, 0.2 mM isopropyl-β-D-thiogalactopyranoside (IPTG) was added to induce protein expression overnight at 18 °C. The following protein purification was performed at 4 °C.
Thirty liters of cells were harvested by centrifugation at 9000× g in a JLA-8.1000 rotor (Beckman Coulter), resuspended in Buffer A (40 mM HEPES-KOH (pH 7.4), 60 mM NaCl and 8% glycerol) supplemented with 1 mM PMSF, 1% (v/v) protease inhibitor cocktail (Mei5bio) and lysed by a French press (JNBIO). Unbroken bacterial cells and large debris were removed by low-speed centrifugation at 10,000× g (rotor JA-25.50, Beckman Coulter) for 1 h and membranes were pelleted by ultra-centrifugation at 160,000× g (rotor 70Ti, Beckman Coulter) for 2 h. All pelleted membranes were solubilized with Buffer B (Buffer A supplemented with 1% (w/v) n-dodecyl-β-d-maltoside (DDM, Anatrace)) for 2 h, and supernatants were collected by ultra-centrifugation (160,000× g for 1 h).
The supernatants were incubated with anti-Flag M2 affinity resin (Sigma Aldrich) with gentle mixing at 4 °C for about 3 h. Resins were recovered by centrifugation at 1000× g (Eppendorf centrifuge 5424 R) and washed extensively with Buffer C (Buffer A supplemented with 0.02% (w/v) DDM). Complexes were then eluted with Buffer D (Buffer A supplemented with 0.02% (w/v) DDM and 0.2 mg/mL 3× Flag peptide). The eluted samples were loaded on a 15%–35% glycerol density gradient and ultra-centrifuged at 130,000× g for 18 h (SW41 rotor, Beckman Coulter). All fractions of the glycerol gradient were manually collected, analyzed with 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie brilliant blue. Fractions with highest homogeneity verified by negative staining electron microscopy were pooled, dialyzed into Buffer C and concentrated to ~18 mg/mL with a 100-kDa-cutoff spin concentrator (Millipore) and stored at −80 °C.
Pull-down assay
HflK–HflC–FtsH complexes containing mutants of FtsH, HflK and HflC were similarly overexpressed in E. coli BL21 (DE3) cells. For each sample, 0.3 L cells were pelleted, lysed and centrifuged at 12,000× g (Eppendorf centrifuge 5424 R) to collect supernatants. Supernatants (normalized by the total protein) were solubilized by 1% (w/v) of DDM (Anatrace). After 1 h extraction, supernatants were incubated with anti-Flag M2 affinity resin (Sigma Aldrich) and the resins were washed extensively with Buffer C and eluted with Buffer D. The eluted samples were subjected to SDS-PAGE (levels of HflC in each sample were used as the loading control) and stained by Coomassie brilliant blue.
In vitro proteolysis assay
A super-fold GFP tag (sfGFP) was added to the N-terminus of CII and the C-terminus of SecY. Amplified genes were constructed into pET28a vectors. For purpose of purification, an 8× His tag was added to the N-terminus of sfGFP-CII and a twin-strep tag to the C-terminus of SecY-sfGFP.
The recombinant pET28a (N-8×HissfGFP-cII) was transformed into E. coli BL21 (DE3) cells for expression. When cells grew to a density of 0.6 at 600 nm at 37 °C, 0.5 mM IPTG was added to induce protein expression at 37 °C for 4 h. Cell pellets from 2 L cultures were re-suspended in lysis buffer containing 20 mM Tris-HCI (pH 8.0) and 150 mM NaCl, lysed by sonication (Sonicators, Qsonica), and centrifuged at 12,000× g for 1 h (rotor JA-25.50, Beckman Coulter). Harvested supernatants were applied onto Ni Smart Beads 6FF (Smart Lifesciences), washed with lysis buffer containing 10 mM imidazole and eluted with lysis buffer containing 250 mM imidazole.
The recombinant pET28a (secY-sfGFPC-twin-strep) was transformed into E. coil C43 (DE3) competent cells to produce un-assembled SecY. When cells grew to a density of 1.0 at 600 nm at 37 °C, 0.2 mM IPTG was added to induce protein expression overnight at 18 °C. Cell pellets from 6 L cultures were re-suspended in the buffer containing 40 mM HEPES-KOH (pH 7.4), 60 mM NaCl, and 8% glycerol, followed by cell lysis of sonication (Sonicators, Qsonica). Cell debris were removed by centrifugation at 12,000× g for 1 h (rotor JA-25.50, Beckman Coulter). Harvested supernatants were further ultra-centrifuged at 160,000× g for 2 h (rotor 70 Ti, Beckman Coulter). Pelleted membrane fractions were solubilized by 1% (w/v) DDM (Anatrace) for 2 h, and supernatants were collected by ultra-centrifugation at 160,000× g for 1 h (70Ti, Beckman Coulter). Collected supernatants were applied to Strep-Tactin® resin (IBA), washed extensively and eluted with buffer containing 40 mM HEPES-KOH (pH 7.4), 60 mM NaCl, 8% glycerol, 0.02% (w/v) DDM and 10 mM desthiobiotin (IBA). The eluted samples were further separated by Superdex Increase 200 column (GE Healthcare).
Proteolysis assay was performed at 37 °C in a PCR machine (Dongsheng Biotech). A standard reaction system contained 40 mM HEPES-KOH (pH 7.4), 60 mM NaCl, 8% glycerol, 0.02% (w/v) DDM, 5 mM magnesium chloride, 0.025 mM zinc acetate and 5 mM ATP. Twelve nanograms of purified HflK–HflC–FtsH complex was mixed with 1.2 ng N-8×HissfGFP-CII or 1.2 ng SecY-sfGFPC-twin-strep. The reaction system was sampled in an interval of 10 min for N-8×HissfGFP-CII or 2 h for SecY-sfGFPC-twin-strep and stopped by adding 5× SDS-loading dye. All the samples were analyzed with 12% SDS-PAGE, visualized by ChemiDocTM MP Imaging system (Bio Rad) and then stained with Coomassie brilliant blue.
Electron microscopy
The sample homogeneity was examined by negative staining electron microscopy. Four microliters of samples (~0.05 mg/mL) was applied onto copper grids and stained with 2% uranyl acetate. The grids were examined with an FEI Tecnai T20 TEM at 120 kV.
Cryo-EM grids were prepared with holey-carbon gold grids (Quantifoil, R1.2/1.3). The grids were glow-discharged for 30 s with a plasma cleaner prior to sample freezing. Four microliters of freshly prepared KCF samples (10 mg/mL) was applied to the grids mounted in the chamber of an FEI Vitrobot Mark IV (4 °C, 100% humidity), and flash-frozen in liquid ethane. Grids were screened with an FEI Talos Arctica (FEI CETA camera) operated at 200 kV. Images were collected with a 300 kV FEI Titan Krios TEM (Gatan K2 summit camera, with GIF energy filter). The movies were acquired using SerialEM80 at a dose rate of 5 e−/s/Å2 and an exposure time of 12 s with 40 frames. The data was collected at the magnification of 130,000× corresponding to a physical pixel size of 1.057 Å/pixel and with defocus ranging from −1.5 to −2.5 μm. Four datasets of images (25,757 in total) were collected separately.
Image processing
The motion correction and electron-dose weighting of the micrographs were performed with MotionCor281 and the contrast transfer function (CTF) parameters were estimated with the program of Gctf.82 Particle picking, classification and structural refinement were done with RELION (3.0 and 3.1 versions).83 For particles auto picking, 2D references were generated from manually picked particles. The initial 3D model was generated using RELION3.0 from 13,607 selected particles.
The four datasets were separately processed as summarized in Supplementary information, Fig. S3. For each dataset, the autopicked particles were first subjected to several rounds of reference-free 2D classification. Particles from good classes were picked and subjected to the first round of 3D classification. The resulting classes without apparent structural deformation were subjected to another round of 3D classification or refinement. The last round of 3D refinement was performed with C4 symmetry imposed. A final set of particles, containing 84,693 particles from four datasets, were pooled together and subjected to 3D refinement with C4 symmetry imposed, resulting in a density map at 3.43 Å (Gold-standard FSC 0.143). Application of a soft mask excluding detergent regions during 3D refinement could improve the overall resolution to 3.27 Å. To further improve the densities of transmembrane regions, particle symmetry expansion (C4 symmetry) and a subsequent mask-based 3D classification (1/4 size of the KCF complex, with “skip alignment” option) were performed using RELION. A total of 274,457 expanded particles were selected for the last round of 3D refinement, resulting in a density map at 3.43 Å. Application of a soft mask only including the SPFH1 domains and the transmembrane helices further improved the densities in these regions, resulting in a map at 3.7 Å (Supplementary information, Fig. S5i, j), which enabled the atomic modeling of the membrane domains.
Model building and refinement
HflK and HflC were modeled de novo using Coot.84 The amino acid sequences of HflK and HflC were subjected to secondary structure prediction using PSIPRED.85 A poly-alanine model was built using Coot, and sequence substitution was performed using bulky side-chain residues as makers. Models of other HflK and HflC conformers were derived by rigid body fitting of the first two models in the density map followed by manual adjustment using Coot. The atomic model of E. coli FtsH periplasmic domain (PDB 4V0B) was docked into the map and manually adjusted.
Model refinement was done with the 1/4 map containing three HflK subunits, three HflC subunits and six FtsH periplasmic domains. Model refinement was performed using real-space refinement (phenix.real_space_refine) in Phenix.86 Model validation (Supplementary information, Table S1) was calculated by MolProbity.87 Figure preparation and structure analysis were performed with PyMOL (pymol.org) and UCSF Chimera.88
Data availability
The cryo-EM map and atomic model of the HflK–HflC–FtsH complex have been deposited in the EMDB and PDB with accession codes EMDB-32002 and 7VHP, respectively. The map and model of a quarter of the complex used for symmetry expansion have been deposited in the EMDB and PDB with accession codes EMDB-32003 and 7VHQ, respectively. All data needed to support the conclusions in this study are included in the main text and supplementary materials.
References
Simons, K. & Ikonen, E. Functional rafts in cell membranes. Nature 387, 569–572 (1997).
Lingwood, D. & Simons, K. Lipid rafts as a membrane-organizing principle. Science 327, 46–50 (2010).
Sezgin, E., Levental, I., Mayor, S. & Eggeling, C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 18, 361–374 (2017).
Lopez, D. & Kolter, R. Functional microdomains in bacterial membranes. Genes Dev. 24, 1893–1902 (2010).
Lucena, D., Mauri, M., Schmidt, F., Eckhardt, B. & Graumann, P. L. Microdomain formation is a general property of bacterial membrane proteins and induces heterogeneity of diffusion patterns. BMC Biol. 16, 97 (2018).
Lopez, D. & Koch, G. Exploring functional membrane microdomains in bacteria: an overview. Curr. Opin. Microbiol. 36, 76–84 (2017).
Browman, D. T., Hoegg, M. B. & Robbins, S. M. The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol. 17, 394–402 (2007).
Yokoyama, H. & Matsui, I. The lipid raft markers stomatin, prohibitin, flotillin, and HflK/C (SPFH)-domain proteins form an operon with NfeD proteins and function with apolar polyisoprenoid lipids. Crit. Rev. Microbiol. 46, 38–48 (2020).
Lapatsina, L., Brand, J., Poole, K., Daumke, O. & Lewin, G. R. Stomatin-domain proteins. Eur. J. Cell Biol. 91, 240–245 (2012).
Danek, M., Valentova, O. & Martinec, J. Flotillins, Erlins, and HIRs: from animal base camp to plant new horizons. Crit. Rev. Plant Sci. 35, 191–214 (2016).
Langhorst, M. F., Reuter, A. & Stuermer, C. A. Scaffolding microdomains and beyond: the function of reggie/flotillin proteins. Cell. Mol. Life Sci. 62, 2228–2240 (2005).
Glebov, O. O., Bright, N. A. & Nichols, B. J. Flotillin-1 defines a clathrin-independent endocytic pathway in mammalian cells. Nat. Cell Biol. 8, 46–54 (2006).
Babuke, T. & Tikkanen, R. Dissecting the molecular function of reggie/flotillin proteins. Eur. J. Cell Biol. 86, 525–532 (2007).
Otto, G. P. & Nichols, B. J. The roles of flotillin microdomains-endocytosis and beyond. J. Cell Sci. 124, 3933–3940 (2011).
Bramkamp, M. & Lopez, D. Exploring the existence of lipid rafts in bacteria. Microbiol. Mol. Biol. Rev. 79, 81–100 (2015).
Garcia-Fernandez, E. et al. Membrane microdomain disassembly inhibits MRSA antibiotic resistance. Cell 171, 1354–1367.e20 (2017).
Wetzel, C. et al. A stomatin-domain protein essential for touch sensation in the mouse. Nature 445, 206–209 (2007).
Huang, M., Gu, G., Ferguson, E. L. & Chalfie, M. A stomatin-like protein necessary for mechanosensation in C. elegans. Nature 378, 292–295 (1995).
Gillespie, P. G. & Walker, R. G. Molecular basis of mechanosensory transduction. Nature 413, 194–202 (2001).
Browman, D. T., Resek, M. E., Zajchowski, L. D. & Robbins, S. M. Erlin-1 and erlin-2 are novel members of the prohibitin family of proteins that define lipid-raft-like domains of the ER. J. Cell Sci. 119, 3149–3160 (2006).
Pearce, M. M., Wormer, D. B., Wilkens, S. & Wojcikiewicz, R. J. An endoplasmic reticulum (ER) membrane complex composed of SPFH1 and SPFH2 mediates the ER-associated degradation of inositol 1,4,5-trisphosphate receptors. J. Biol. Chem. 284, 10433–10445 (2009).
Lu, J. P., Wang, Y., Sliter, D. A., Pearce, M. M. & Wojcikiewicz, R. J. RNF170 protein, an endoplasmic reticulum membrane ubiquitin ligase, mediates inositol 1,4,5-trisphosphate receptor ubiquitination and degradation. J. Biol. Chem. 286, 24426–24433 (2011).
Ikonen, E., Fiedler, K., Parton, R. G. & Simons, K. Prohibitin, an antiproliferative protein, is localized to mitochondria. FEBS Lett. 358, 273–277 (1995).
Steglich, G., Neupert, W. & Langer, T. Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria. Mol. Cell. Biol. 19, 3435–3442 (1999).
McClung, J. K. et al. Isolation of a cDNA that hybrid selects antiproliferative mRNA from rat liver. Biochem. Biophys. Res. Commun. 164, 1316–1322 (1989).
Artal-Sanz, M. & Tavernarakis, N. Prohibitin and mitochondrial biology. Trends Endocrinol. Metab. 20, 394–401 (2009).
Merkwirth, C. & Langer, T. Prohibitin function within mitochondria: essential roles for cell proliferation and cristae morphogenesis. Biochim. Biophys. Acta 1793, 27–32 (2009).
Wang, D. et al. Prohibitin ligands: a growing armamentarium to tackle cancers, osteoporosis, inflammatory, cardiac and neurological diseases. Cell. Mol. Life Sci. 77, 3525–3546 (2020).
Kihara, A., Akiyama, Y. & Ito, K. A protease complex in the Escherichia coli plasma membrane: HflKC (HflA) forms a complex with FtsH (HflB), regulating its proteolytic activity against SecY. EMBO J. 15, 6122–6131 (1996).
Glynn, S. E. Multifunctional mitochondrial AAA proteases. Front. Mol. Biosci. 4, 34 (2017).
Patron, M., Sprenger, H. G. & Langer, T. m-AAA proteases, mitochondrial calcium homeostasis and neurodegeneration. Cell Res. 28, 296–306 (2018).
Arnold, I. & Langer, T. Membrane protein degradation by AAA proteases in mitochondria. Biochim. Biophys. Acta 1592, 89–96 (2002).
Quiros, P. M., Langer, T. & Lopez-Otin, C. New roles for mitochondrial proteases in health, ageing and disease. Nat. Rev. Mol. Cell Biol. 16, 345–359 (2015).
Bohovych, I., Chan, S. S. & Khalimonchuk, O. Mitochondrial protein quality control: the mechanisms guarding mitochondrial health. Antioxid. Redox Signal. 22, 977–994 (2015).
Brand, J. et al. A stomatin dimer modulates the activity of acid-sensing ion channels. EMBO J. 31, 3635–3646 (2012).
Yokoyama, H., Fujii, S. & Matsui, I. Crystal structure of a core domain of stomatin from Pyrococcus horikoshii Illustrates a novel trimeric and coiled-coil fold. J. Mol. Biol. 376, 868–878 (2008).
Kuwahara, Y. et al. Unusual thermal disassembly of the SPFH domain oligomer from Pyrococcus horikoshii. Biophys. J. 97, 2034–2043 (2009).
Takekawa, N. et al. Structure of Vibrio FliL, a new stomatin-like protein that assists the bacterial flagellar motor function. mBio 10, e00292–19 (2019).
Yoshinaka, T. et al. Structural basis of mitochondrial scaffolds by prohibitin complexes: insight into a role of the coiled-coil region. iScience 19, 1065–1078 (2019).
Tatsuta, T., Model, K. & Langer, T. Formation of membrane-bound ring complexes by prohibitins in mitochondria. Mol. Biol. Cell 16, 248–259 (2005).
Boehm, M. et al. Structural and mutational analysis of band 7 proteins in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 191, 6425–6435 (2009).
Kowalski, M. P. et al. Host resistance to lung infection mediated by major vault protein in epithelial cells. Science 317, 130–132 (2007).
Tanaka, H. et al. The structure of rat liver vault at 3.5 angstrom resolution. Science 323, 384–388 (2009).
Back, J. W. et al. A structure for the yeast prohibitin complex: Structure prediction and evidence from chemical crosslinking and mass spectrometry. Protein Sci. 11, 2471–2478 (2002).
Briere, L. A. K. & Dunn, S. D. The periplasmic domains of Escherichia coli HflKC oligomerize through right-handed coiled-coil interactions. Biochemistry 45, 8607–8616 (2006).
Saikawa, N., Akiyama, Y. & Ito, K. FtsH exists as an exceptionally large complex containing HflKC in the plasma membrane of Escherichia coli. J. Struct. Biol. 146, 123–129 (2004).
Kihara, A., Akiyama, Y. & Ito, K. Host regulation of lysogenic decision in bacteriophage lambda: transmembrane modulation of FtsH (HflB), the cII degrading protease, by HflKC (HflA). Proc. Natl. Acad. Sci. USA 94, 5544–5549 (1997).
Ito, K. & Akiyama, Y. Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 59, 211–231 (2005).
Tomoyasu, T. et al. Topology and subcellular localization of FtsH protein in Escherichia coli. J. Bacteriol. 175, 1352–1357 (1993).
Scharfenberg, F. et al. Structure and evolution of N-domains in AAA metalloproteases. J. Mol. Biol. 427, 910–923 (2015).
Boute, N. et al. NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat. Genet. 24, 349–354 (2000).
Bouchireb, K. et al. NPHS2 mutations in steroid-resistant nephrotic syndrome: a mutation update and the associated phenotypic spectrum. Hum. Mutat. 35, 178–186 (2014).
Langklotz, S., Baumann, U. & Narberhaus, F. Structure and function of the bacterial AAA protease FtsH. Biochim. Biophys. Acta 1823, 40–48 (2012).
Tomoyasu, T. et al. Escherichia coli Ftsh is a membrane-bound, Atp-dependent protease which degrades the heat-shock transcription factor sigma 32. EMBO J. 14, 2551–2560 (1995).
Ogura, T. et al. Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol. Microbiol. 31, 833–844 (1999).
Shotland, Y. et al. Proteolysis of the phage lambda CII regulatory protein by FtsH (HflB) of Escherichia coli. Mol. Microbiol. 24, 1303–1310 (1997).
Leonhard, K. et al. AAA proteases with catalytic sites on opposite membrane surfaces comprise a proteolytic system for the ATP-dependent degradation of inner membrane proteins in mitochondria. EMBO J. 15, 4218–4229 (1996).
Di Bella, D. et al. Mutations in the mitochondrial protease gene AFG3L2 cause dominant hereditary ataxia SCA28. Nat. Genet. 42, 313–321 (2010).
Pierson, T. M. et al. Whole-exome sequencing identifies homozygous AFG3L2 mutations in a spastic ataxia-neuropathy syndrome linked to mitochondrial m-AAA proteases. PLoS Genet. 7, e1002325 (2011).
Casari, G. et al. Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell 93, 973–983 (1998).
Akiyama, Y., Kihara, A., Mori, H., Ogura, T. & Ito, K. Roles of the periplasmic domain of Escherichia coli FtsH (HflB) in protein interactions and activity modulation. J. Biol. Chem. 273, 22326–22333 (1998).
Kihara, A., Akiyama, Y. & Ito, K. FtsH is required for proteolytic elimination of uncomplexed forms of SecY, an essential protein translocase subunit. Proc. Natl. Acad. Sci. USA 92, 4532–4536 (1995).
Akiyama, Y., Kihara, A. & Ito, K. Subunit a of proton ATPase F-0 sector is a substrate of the FtsH protease in Escherichia coli. FEBS Lett. 399, 26–28 (1996).
Kohanski, M. A., Dwyer, D. J., Wierzbowski, J., Cottarel, G. & Collins, J. J. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 135, 679–690 (2008).
Nonaka, G., Blankschien, M., Herman, C., Gross, C. A. & Rhodius, V. A. Regulon and promoter analysis of the E. coli heat-shock factor, sigma32, reveals a multifaceted cellular response to heat stress. Genes Dev. 20, 1776–1789 (2006).
Letunic, I., Khedkar, S. & Bork, P. SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460 (2021).
Rivera-Milla, E., Stuermer, C. A. & Malaga-Trillo, E. Ancient origin of reggie (flotillin), reggie-like, and other lipid-raft proteins: convergent evolution of the SPFH domain. Cell. Mol. Life Sci. 63, 343–357 (2006).
Hinderhofer, M. et al. Evolution of prokaryotic SPFH proteins. BMC Evol. Biol. 9, 10 (2009).
Morrow, I. C. et al. Flotillin-1/reggie-2 traffics to surface raft domains via a novel golgi-independent pathway. Identification of a novel membrane targeting domain and a role for palmitoylation. J. Biol. Chem. 277, 48834–48841 (2002).
Neumann-Giesen, C. et al. Membrane and raft association of reggie-1/flotillin-2: role of myristoylation, palmitoylation and oligomerization and induction of filopodia by overexpression. Biochem. J. 378, 509–518 (2004).
Wai, T. et al. The membrane scaffold SLP2 anchors a proteolytic hub in mitochondria containing PARL and the i-AAA protease YME1L. EMBO Rep. 17, 1844–1856 (2016).
Chiba, S., Ito, K. & Akiyama, Y. The Escherichia coli plasma membrane contains two PHB (prohibitin homology) domain protein complexes of opposite orientations. Mol. Microbiol. 60, 448–457 (2006).
Pike, L. J. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res. 47, 1597–1598 (2006).
Pralle, A., Keller, P., Florin, E. L., Simons, K. & Horber, J. K. H. Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J. Cell Biol. 148, 997–1007 (2000).
Rungaldier, S. et al. Structure-function analysis of human stomatin: a mutation study. PLoS ONE 12, e0178646 (2017).
Huber, T. B. et al. Podocin and MEC-2 bind cholesterol to regulate the activity of associated ion channels. Proc. Natl. Acad. Sci. USA 103, 17079–17086 (2006).
Lingwood, D., Kaiser, H. J., Levental, I. & Simons, K. Lipid rafts as functional heterogeneity in cell membranes. Biochem. Soc. Trans. 37, 955–960 (2009).
Osman, C., Merkwirth, C. & Langer, T. Prohibitins and the functional compartmentalization of mitochondrial membranes. J. Cell Sci. 122, 3823–3830 (2009).
Price, M. P., Thompson, R. J., Eshcol, J. O., Wemmie, J. A. & Benson, C. J. Stomatin modulates gating of acid-sensing ion channels. J. Biol. Chem. 279, 53886–53891 (2004).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Buchan, D. W., Minneci, F., Nugent, T. C., Bryson, K. & Jones, D. T. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res. 41, W349–W357 (2013).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Pettersen, E. F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Acknowledgements
We thank the Core Facilities at the School of Life Sciences, Peking University for help with negative staining EM; the Cryo-EM Platform and the Electron Microscopy Laboratory of Peking University for help with data collection; the High-performance Computing Platform of Peking University for help with computation; and the National Center for Protein Sciences at Peking University for assistance in key experiments. The work was supported by the National Natural Science Foundation of China (31725007 and 31630087 to N.G.; 31922036 to N.L.; 31800625 to C.M.), the National Key R&D Program of China (2019YFA0508904 to N.G.), the Qidong-SLS Innovation Fund (to N.G.), and the China Postdoctoral Science Foundation (2018M631249 and 2021T140016 to C.M.). C.M. and C.W. were supported in part by the Postdoctoral Fellowship of Peking-Tsinghua Center for Life Sciences.
Author information
Authors and Affiliations
Contributions
N.G. designed and supervised the project; C.W. purified the sample with help from D.L., L.Y. and W.Y. and performed protease and pull-down assays; C.M. processed cryo-EM data with help from N.L.; N.G. built atomic models and analyzed the structure with C.M. and C.W.; C.M. prepared figures; N.G. wrote the manuscript; C.W. and C.M. revised the manuscript. All the authors approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Ma, C., Wang, C., Luo, D. et al. Structural insights into the membrane microdomain organization by SPFH family proteins. Cell Res 32, 176–189 (2022). https://doi.org/10.1038/s41422-021-00598-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41422-021-00598-3