Abstract
Triple-negative breast cancer (TNBC) is an aggressive type of cancer associated with a poor prognosis. Identification of novel therapeutic targets in TNBC is urgently needed. Here, we investigated the microRNA (miRNA) expression signature of TNBC using clinical specimens. In total, 104 miRNAs (56 upregulated and 48 downregulated) were significantly dysregulated in TNBC tissues; miR-204-5p showed the most dramatic downregulation. We then examined the antitumor roles of miR-204-5p in breast cancer (BC) cells. Notably, cancer cell migration and invasion were significantly reduced by ectopic expression of miR-204-5p in BC cells. Genome-wide gene expression analysis and in silico database search revealed that 32 genes were putative miR-204-5p targets. High expression of AP1S3, RACGAP1, ELOVL6, and LRRC59 was significantly associated with poor prognosis in patients with BC, and adaptor-related protein complex 1 sigma 3 subunit (AP1S3) was directly regulated by miR-204-5p, as demonstrated by luciferase reporter assays. AP1S3 overexpression was detected in TNBC clinical specimens and enhanced cancer cell aggressiveness. We further analyzed downstream RNA networks regulated by AP1S3 in BC cells. Overall, this miRNA signature is expected to be an effective tool for identification of miRNA-mediated molecular mechanisms of TNBC pathogenesis.
Similar content being viewed by others
Introduction
Despite advances in early detection systems and currently developed molecular targeted therapies, breast cancer (BC) is a leading cause of cancer-related death among women in industrialized countries [1, 2]. Based on the St. Gallen Consensus Meeting of Breast Cancer, BC can be classified into five subtypes as follows: luminal A-like type, luminal B-like type, hormone receptor-positive and human epidermal growth factor receptor 2 (HER2)-positive type, hormone receptor-negative and HER2-postive type, and triple-negative type [3,4,5]. Triple-negative BC (TNBC) includes tumors without the expression of estrogen receptor (ER), progesterone receptor (PR), and HER2 and largely overlaps with the basal subtype, representing only 15–20% of BC cases [3,4,5]. Due to the aggressive nature of TNBC and the lack of novel targeted therapies, TNBC has a significantly worse prognosis than other types of BC [6, 7]. Recent genomic approaches have offered the possibility to improve treatment approaches through the identification of novel targets for TNBC anticancer therapy.
MicroRNAs (miRNAs) are small noncoding RNAs that fine-tune the expression of protein coding/noncoding RNAs by repressing translation or cleaving RNA transcripts in a sequence-dependent manner [8, 9]. Many studies have shown that miRNAs play important roles in various physiological processes, such as cell differentiation, proliferation, and apoptosis [10, 11]. In humans, dysregulation of miRNAs is associated with many human diseases, including BC pathogenesis [12,13,14]. Based on the unique nature of miRNA biology, a single miRNA controls many RNA transcripts in normal and diseased cells [9, 15]. Identification of aberrantly expressed miRNAs in cancer cells is the first step for elucidation of miRNA-mediated cancer pathways.
Advanced RNA sequencing technologies are suitable for construction of miRNA expression signatures. We have provided miRNA expression signatures based on RNA sequencing in several cancer types, e.g., bladder cancer, castration-resistant prostate cancer, pancreatic cancer, and head and neck cancer [16,17,18,19]. In this study, we created the miRNA expression signature of TNBC by RNA sequencing using clinical specimens and found that 104 miRNAs (56 upregulated and 48 downregulated) were significantly dysregulated in TNBC tissues. These dysregulated miRNAs represent an important starting point for elucidation of the molecular pathology of TNBC.
Our signature showed that miR-204-5p was significantly downregulated in TNBC tissues. Therefore, we aimed to identify targets of miR-204-5p involved in TNBC pathogenesis. Identification of aberrantly expressed miRNA-mediated cancer networks is expected to provide insights into the potential mechanisms underlying TNBC pathogenesis and to facilitate the establishment of novel therapeutic targets.
Materials and methods
Patients, BC specimens, and cell lines
Thirty-five clinical tissue specimens were collected from patients with BC who underwent surgical resection at Gunma University Hospital and Kagoshima University Hospital between 2003 and 2018. The clinicopathological features of patients with BC are shown in Supplementary Table 1. All patients in this study provided informed consent, and the study protocol was approved by the Institutional Review Board of Gunma University (approval nos. 2016-023 and 2017-163) and Kagoshima University (approval no. 28-65).
MDA-MB-231 and MDA-MB-157 cells were used in this study. All cell lines were obtained from Public Health England (Salisbury, UK).
Total RNA, including miRNA, was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA).
Construction of the miRNA expression signature for TNBC
Small RNA sequencing was performed using a HiSeq 2000 instrument (Illumina, San Diego, CA, USA) for five TNBC samples and three normal breast epithelial tissue samples (Supplementary Table 1). Small RNA sequencing and data mining procedures were described in our previous studies [16,17,18,19,20].
Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR)
The procedure for qRT-PCR has been described previously [21,22,23,24]. Briefly, the expression levels of miRNAs were analyzed by TaqMan qRT-PCR assays (miR-204-5p, assay ID: 000508; Applied Biosystems, Foster City, CA, USA). Data were normalized to RNU48 (assay ID: 001006; Applied Biosystems). In addition, the expression levels of target genes of miR-204-5p were assessed with the following TaqMan probes and primers: AP1S3, assay ID: Hs00950999; Applied Biosystems, normalized to GUSB (assay ID: Hs00939627_ml; Applied Biosystems).
Effects of transfection with miRNA mimic or small-interfering RNA (siRNA) on cell proliferation, migration, and invasion
The transfection procedures for miRNAs or siRNAs into cancer cells were described previously [21,22,23,24]. The following mature miRNAs or siRNAs were used in this study: mature miRNAs (Ambion Pre-miR miRNA precursor; miR-204-5p, ID: PM11116; Applied Biosystems), siRNAs (Stealth Select RNAi siRNA; si-AP1S3, ID: HSS134239 and HSS134240; Invitrogen), and negative control miRNA/siRNA (product ID: AM17010; Thermo Fisher Scientific, Waltham, MA, USA).
Protocols for determining cell proliferation, migration, and invasion were described previously [21,22,23,24].
Genome-wide gene expression and in silico analyses for the identification of genes regulated by miR-204-5p in BC cells
Microarray gene expression analysis and in silico analysis were used to identify putative target genes regulated by miR-204-5p. The microarray data of miR-204-5p transfectants were deposited in the Gene Expression Omnibus (GEO) repository under accession number GSE115801. Gene expression data by microarray analyses of TNBC clinical specimens were deposited in the GEO database (GSE118539). Putative target genes having a binding site for miR-204-5p were detected by TargetScan Human ver.7.1 (http://www.targetscan.org/vert_71/). Our strategy for identification of miR-204-5p target genes is outlined in Supplementary Fig. 1.
The Cancer Genome Atlas (TCGA) data analysis
To assess the clinical significance of miR-204-5p-targeted genes, we used the RNA sequencing database in TCGA. High and low expression were defined by dividing the clinical data population in half based on the expression level. Gene expression and clinical data were obtained from cBioportal and OncoLnc (data downloaded November 1, 2017). Data mining procedures were described in our previous studies [21,22,23,24].
Plasmid construction and dual-luciferase reporter assays
The following sequences were inserted into the psiCHECk-2 vector (C8021; Promega, Madison, WI, USA): the wild-type sequence of the 3′-untranslated region (UTR) of AP1S3 or the deletion-type, which lacked the miR-204-5p binding sites (positions 111−117 or 1993–2000; Fig. 2c, upper panel).
The cloned vectors were cotransfected into MDA-MB-231 cells with mature miR-204-5p. The procedures for transfection and dual-luciferase reporter assays were described in previous studies [21,22,23,24].
Western blotting and immunohistochemistry
Anti-human AP1S3 rabbit polyclonal IgG (1:500; ab205509; Abcam, Cambridge, UK) was used as a primary antibody. Anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mouse monoclonal IgG (1:2000; SAF6698; Wako, Osaka, Japan) was used as an internal control. The protocol for western blot analysis was described previously [21,22,23,24].
Anti-human AP1S3 rabbit polyclonal IgG (1:200; PA5-23894; Thermo Fisher Scientific) was used as a primary antibody for immunohistochemistry. The procedures were described in previous studies [21,22,23,24].
Identification of downstream genes regulated by AP1S3 in BC cells
The microarray data for si-AP1S3 transfectants were deposited in the GEO repository under accession number GSE115909.
Statistical analysis
Mann−Whitney U tests were used to analyze differences between two groups. Bonferroni-adjusted Mann−Whitney U tests were used to analyze the differences among more than three groups. Statistical tests were performed using Expert StatView software (version 5.0, SAS Institute Inc., Cary, NC, USA). Survival analysis was performed using the Kaplan–Meier method, log-rank tests, and multivariable Cox hazard regression analyses with JMP software (version 13; SAS Institute Inc., Cary, NC, USA).
Results
Small RNA sequencing of TNBC specimens and construction of miRNA signatures
To create the miRNA expression signature of TNBC, we performed small RNA sequencing of eight RNA libraries (derived from three normal breast epithelial tissues and five TNBC tissues). The clinical features of clinical specimens are summarized in Supplementary Table 1. We obtained between 10,112,225 and 14,568,843 reads in this analysis. After filing and trimming of the sequenced reads, between 7,076,489 and 12,271,020 miRNA reads (larger than 19 nucleotides) were mapped in the human genome (Supplementary Table 2). Human genome-matched sequenced reads were divided into small RNAs according to their biological features (Supplementary Table 2).
We then created the miRNA expression signature that included miRNAs exhibiting significant up- or downregulation (false discovery rate < 0.05) in TNBC tissues (Table 1). In total, 56 upregulated miRNAs and 48 downregulated miRNAs were detected from aligned reads using R program (Table 1). Among these dysregulated miRNAs, 23 were annotated as passenger strands of miRNAs in the miRBase database (Release 22, http://www.mirbase.org/).
Expression levels of miR-204-5p in BC specimens and cell lines
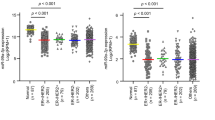
To confirm the miRNA expression signature, expression levels of miR-204-5p in normal breast epithelial tissues (n = 11), TNBC tissues (n = 16), and cell lines (MDA-MB-231 and MDA-MB-157) were evaluated. The expression level of miR-204-5p was significantly downregulated in TNBC specimens (Fig. 1a). Additionally, the expression levels of this miRNA in the two cell lines were lower than those in normal BC epithelial specimens (Fig. 1a).
Effects of ectopic expression of miR-204-5p on BC cells. a Expression levels of miR-204-5p in TNBC clinical specimens and cell lines (MDA-MB-231 and MDA-MB-157). RNU48 was used as an internal control. b Cell proliferation was determined by XTT assays 72 h after transfection with miR-204-5p p. *p < 0.05. c Results of cell migration assays. *p < 0.05. d Cell invasion activity was determined using Matrigel invasion assays. *p < 0.05
The clinical features of patients with TNBC are summarized in Supplementary Table 1.
Effects of restoring miR-204-5p on cell proliferation, migration, and invasion in BC cells
To evaluate the antitumor effects of miR-204-5p, we applied gain-of-function assays in TNBC cell lines (MDA-MB-231 and MDA-MB-157). Ectopic expression of miR-204-5p significantly blocked cancer cell proliferation, migration, and invasion (Fig. 1b–d).
Identification of putative target genes regulated by miR-204-5p in BC cells and their clinical significance
To identify putative oncogenic genes regulated by miR-204-5p in TNBC cells, we applied three different datasets: (1) candidates of miR-204-5p binding genes in the TargetScan database, (2) genes downregulated by miR-204-5p transfection in MDA-MB-231 cells (GEO accession number, GSE115801), and (3) our original gene expression data from TNBC clinical specimens. Our strategy for identification of putative target genes regulated by miR-204-5p in TNBC cells is shown in Supplementary Fig. 1. Finally, 32 genes were identified as miR-204-5p-regulated oncogenes (Table 2). Oligo microarrays (Human GE 60K; Agilent Technologies) were used for gene expression analyses of TNBC clinical specimens. The microarray data were deposited into GEO (https://www.ncbi.nlm.nih.gov/geo/), with accession number GSE118539.
Next, to investigate the clinical significance of these targets, we analyzed the relationships between gene expression and prognosis in patients with BC using TCGA database. Among these targets, high expression levels of four genes (AP1S3: p = 0.00823, RACGAP1: p = 0.0277, ELOVL6: p = 0.0448, and LRRC59: p = 0.0456) were significantly associated with poor prognosis in patients with BC by TCGA database analyses (Fig. 4c and Supplementary Fig. 2). Expression levels of three genes (RACGAP1, ELOVL6, and LRRC59) were decreased by transfected with miR-204-5p into BC cells (Supplementary Fig. 2).
Direct regulation of AP1S3 by miR-204-5p in BC cells
We then examined whether miR-204-5p regulated AP1S3 in TNBC cells. We confirmed that gene expression of AP1S3 was significantly decreased in TNBC cells transfected with miR-204-5p (Fig. 2a). Additionally, western blot analyses revealed that AP1S3 protein levels in TNBC cells were decreased by transfection with miR-204-5p (Fig. 2b).
Direct regulation of AP1S3 by miR-204-5p in BC cells. a Expression levels of AP1S3 mRNA 72 h after transfection with 10 nM miR-204-5p into cell line. GUSB was used as an internal control. *p < 0.0001. b Expression of AP1S3 protein 96 h after transfection with miR-204-5p. GAPDH was used as a loading control. c miR-204-5p binding sites in the 3′-untranslated region (3′-UTR) of AP1S3 mRNA. Dual-luciferase reporter assays using vectors encoding putative miR-204-5p target sites (positions 111−117 or 1993−2000) in the AP1S3 3′-UTR for both wild-type and deleted regions. Renilla luciferase values were normalized to firefly luciferase values. *p < 0.005
Next, we performed luciferase reporter assays with MDA-MB-231 cells to determine whether miR-204-5p directly targeted the 3′-UTR of AP1S3. According to the TargetScan human database, the binding sites for miR-204-5p in the 3′-UTR of AP1S3 consisted of two regions (positions 111−117 and 1993−2000; Fig. 2c, upper panel).
Moreover, we showed that miR-204-5p suppressed reporter activity in cells transfected with the wild-type AP1S3 vector compared with that in mock or miR-control transfectants (p < 0.005, Fig. 2c, lower panel), whereas transfectants of the mutant vector were not decreased. These data indicated that miR-204-5p was directly bound to two putative binding sites in the 3′-UTR of AP1S3.
In addition, adaptor protein complex 1 (AP-1) consists of three member of genes, AP1S1, AP1S2, and AP1S3. In this study, we identified AP1S1 and AP1S2 as putative targets of miR-204-5p regulation in BC cells. Expression of AP1S1 was upregulated in TNBC clinical specimens. AP1S2 was not upregulated in TNBC clinical specimens (Supplementary Fig. 3A). Expression of AP1S1 and AP1S2 was reduced by miR-204-5p in BC cells (Supplementary Fig. 3B).
Effects of AP1S3 knockdown on cell proliferation, migration, and invasion in BC cells
To assess the function of AP1S3 in TNBC cells, loss-of-function assays using siRNA were performed. We evaluated the knockdown efficiency of si-AP1S3-transfected TNBC cell lines. Downregulation of AP1S3/AP1S3 was detected in si-AP1S3 transfectants (Fig. 3a, b).
Effects of silencing AP1S3 in BC cells. a AP1S3 mRNA expression 72 h after transfection of BC cells with 10 nM si-AP1S3. GUSB was used as an internal control. *p < 0.0001. b AP1S3 protein expression 96 h after transfection with si-AP1S3. GAPDH was used as a loading control. c Cell proliferation was determined with XTT assays 72 h after transfection with 10 nM si-AP1S3. *p < 0.0001. d Results of cell migration assays. *p < 0.0001. e Results of Matrigel invasion assays. *p < 0.0001
Cancer cell proliferation, migration, and invasion were significantly reduced in si-AP1S3 transfectants compared with those in mock- or miR-control-transfected TNBC cell lines (Fig. 3c–e).
In addition, we investigated the oncogenic roles of AP1S3 by using other cancer cell lines (MDA-MB157: breast cancer and H1299: lung adenocarcinoma). A large number of cohort analyses by TCGA datasets showed that high expression of AP1S3 significantly predicted poor prognosis of lung adenocarcinoma (Supplementary Fig. 4A). Our functional assays showed that cancer cell aggressiveness was markedly suppressed by transfecting with si-AP1S3 into MDA-MB-157 and H1299 cell lines (Supplementary Fig. 4B).
Expression of AP1S3 in BC clinical specimens and its clinical significance
AP1S3 expression levels were significantly upregulated in TNBC tissues (Fig. 4a). Spearman’s rank tests showed a negative correlation between the expression of AP1S3 and miR-204-5p (p = 0.0129, r = −0.488; Fig. 4b). Based on the TCGA database analysis, the overall survival rates of patients with BC were significantly shorter in patients with elevated AP1S3 expression compared with those in patients with low expression (p = 0.00823; Fig. 4c).
Expression of AP1S3 in TNBC clinical specimens. a Expression levels of AP1S3 in TNBC clinical specimens. GUSB was used as an internal control. b Spearman's rank test was used to evaluate the correlations between AP1S3 expression and miR-204-5p. c The 5-year survival rates were analyzed by Kaplan−Meier survival curves and log-rank statistics. d Immunostaining showed that AP1S3 was strongly expressed in cancer lesions, but not in noncancerous epithelial tissues
Finally, we performed univariate and multivariate Cox hazard regression analyses to investigate the clinical significance of AP1S3 expression for overall survival in patients with BC. After multivariate analysis, high AP1S3 expression levels, age (>60 years), lymph node metastasis status, and metastasis status were found to be independent predictive factors for overall survival (hazard ratio (HR) = 1.77, p = 0.0031; HR = 2.02, p = 0.0004; HR = 2.18, p = 0.0002; and HR = 3.19, p = 0.0017, respectively; Supplementary Table 3). These findings showed that overexpression of AP1S3 was relevant to cancer aggressiveness and was associated with poor outcomes.
Further examination of the protein expression levels of AP1S3 in TNBC clinical specimens by immunostaining showed that AP1S3 was strongly expressed in cancer lesions, but not in noncancerous epithelial tissues (Fig. 4d).
Downstream genes affected by silencing of AP1S3 in BC cells
To identity downstream genes regulated by AP1S3, we used genome-wide gene expression data (si-AP1S3-transfected cells and TNBC expression data). In total, 32 genes were identified as putative downstream genes regulated by AP1S3 in BC cells (Supplementary Table 4). Expression data were deposited into GEO (https://www.ncbi.nlm.nih.gov/geo/), with accession number GSE115909.
Discussion
Recent studies associated with the Human Genome Project have revealed that in human cells, various RNAs (protein-coding RNAs, small noncoding RNAs, and large noncoding RNAs) form complicated RNA networks and are responsible for many biological processes [25]. Disruption of tightly regulated RNA networks by aberrantly expressed miRNAs contributes to cancer cell development, metastasis, and drug resistance [26, 27]. Therefore, the identification of miRNAs that are abnormally expressed in cancer cells will facilitate the search for novel RNA networks in cancer cells. To search for miRNAs characteristic of cancer cells, we created miRNA expression signatures using clinical specimens from patients with several types of cancers based on the most advanced genomic strategies [16,17,18,19, 28,29,30].
In this study, we created a novel TNBC miRNA expression signature by RNA sequencing and found that 104 miRNAs (56 upregulated and 48 downregulated) were dysregulated in TNBC tissues. Importantly, some passenger strands of miRNAs were aberrantly expressed in cancer tissues, e.g., miR-202-5p, miR-99a-3p, miR-675-3p, miR-10b-3p, miR-145-3p, and miR-30a-3p. It is generally believed that the passenger strands of miRNAs are degraded and therefore have no function [31, 32]. However, our recent studies have shown that miR-99a-3p and miR-145-3p act as antitumor miRNAs by targeting several oncogenes, including MELK, NCAPG, BUB1, and CDK1, in prostate cancer [17, 33]. The involvement of passenger strands of miRNAs in cancer pathogenesis is a new concept of cancer research. The miRNA signature we obtained in this study represents a major advancement in TNBC research and is expected to facilitate the identification of novel RNA networks associated with the aggressiveness of TNBC.
In the present study, we focused on miR-204-5p because its expression showed the greatest downregulation in TNBC tissues in our signature. Our present data showed that ectopic expression of miR-204-5p inhibited processes associated with cancer cell malignancy, suggesting that this miRNA had antitumor effects in BC cells. Previous studies have demonstrated that miR-204-5p acts as an antitumor miRNA in several cancers [34,35,36,37]. In BC cells, the expression of miR-204-5p has been shown to suppress the epithelial–mesenchymal transition (EMT) phenotype in cancer cells by targeting the homeobox protein sineoculis homeobox homolog 1 (SIX1) [38]. Overexpression of SIX1 was observed in BC and was found to be related to BC cell metastasis by promoting the EMT [39]. Further studies of such miRNA-regulated molecular networks will contribute to the identification of molecular mechanisms mediating carcinogenesis.
We also attempted to identify RNA networks regulated by miR-204-5p in BC cells. Our in silico and genome-wide gene expression analyses revealed 32 candidate oncogenes that may be controlled by miR-204-5p in TNBC cells. Among these targets, four overexpressed genes (AP1S3: p = 0.00823, RACGAP1: p = 0.0277, ELOVL6: p = 0.0448, and LRRC59: p = 0.0456) were predicted to be associated with poor prognosis in patients with BC in TCGA database. Rac GTPase-activating protein 1 (RACGAP1) binds the activated form of Rho GTPase and stimulates GTP hydrolysis, resulting in negative regulation of Rho-mediated signals. Several studies have shown that expression of RACGAP1 is directly correlated with shorter disease-free survival in patients with BC [40,41,42]. Moreover, RACGAP1 was found to behave as an oncogene in BC cells. Functional analysis of these target genes will be helpful for improving our understanding of the molecular pathology of TNBC.
In this study, we focused on AP1S3 because its expression was associated with poor prognosis in patients with BC. In addition, no studies have examined the relationship between AP1S3 and cancer pathogenesis. AP1S3 is a component of adaptor protein complex 1 (AP-1), and three paralogous genes, AP1S1, AP1S2, and AP1S3 exist in the human genome [43]. AP1S proteins are essential for the stability of the AP-1 complex, and mutation of AP1S genes may disrupt the AP-1 complex. AP-1 complexes are involved in clathrin-mediated vesicular transport from the Golgi or endosomes [44]. Moreover, the AP-1 complex has been implicated in the formation of autophagosomes [45]. In a recent study, mutations in the AP1S3 gene were identified in patients with pustular psoriasis, a severe autoinflammatory skin disorder [46, 47]. To the best of our knowledge, this is the first report demonstrating that AP1S3 was directly regulated by antitumor miR-204-5p and that expression of AP1S3 was involved in cancer pathogenesis. Interestingly, other AP1S members, i.e., AP1S1 and AP1S2, were identified as miR-204-5p-regulated targets in TNBC cells. Further studies are needed to determine how aberrant expression of these genes is involved in cancer development.
To investigate the biological significance of AP1S3 in BC cells, we investigated the downstream genes modulated by AP1S3. A total of 33 putative target genes were downregulated by transfecting with si-AP1S3 into BC cells. These genes will be useful for understanding molecular pathogenesis of AP1S3-modulated BC cells. For example, YWHAZ, a member of the 14-3-3 family was high expression of BC clinical specimens and its aberrant expression contributed to chemotherapy resistance and recurrence of BC [48,49,50]. More recently, miR-451 was downregulated in paclitaxel-resistant BC cells and YWHAZ was a direct target of miR-451 in BC cells [51]. Interestingly, our miRNA signature of TNBC showed that miR-451a was significantly downregulated in cancer tissues. Further studies of antitumor miRNA-modulated molecular pathways in TNBC may provide novel insights into the molecular pathogenesis of this disease.
In conclusion, dysregulated miRNAs, including passenger strands of miRNAs, were successfully identified by RNA sequencing-based signatures in TNBC cells. Downregulation of miR-204-5p was detected by our present signature, and this miRNA was found to function as an antitumor miRNA in TNBC cells. Direct regulation of AP1S3 by miR-204-5p was detected in BC cells, and AP1S3 expression was found to be involved in BC pathogenesis. Identification of novel cancer networks mediated by aberrantly expressed miRNAs may improve our understanding of the molecular pathogenesis of TNBC. Moreover, our newly created RNA sequencing-based miRNA signature establishes a basis for further TNBC research.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108.
Koboldt DC, Fulton RS, McLellan MD, Schmidt H, Kalicki-Veizer J, McMichael JF, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70.
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52.
Coates AS, Winer EP, Goldhirsch A, Gelber RD, Gnant M, Piccart-Gebhart M, et al. Tailoring therapies—improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann Oncol. 2015;26:1533–46.
Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thurlimann B, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013;24:2206–23.
Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–48.
Poggio F, Bruzzone M, Ceppi M, Ponde NF, La Valle G, Del Mastro L, et al. Platinum-based neoadjuvant chemotherapy in triple-negative breast cancer: a systematic review and meta-analysis. Ann Oncol. 2018;29:1497–508.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33.
Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40.
He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network—another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7:819–22.
Shenoy A, Blelloch RH. Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nat Rev Mol Cell Biol. 2014;15:565–76.
Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–33.
Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–22.
Kurozumi S, Yamaguchi Y, Kurosumi M, Ohira M, Matsumoto H, Horiguchi J. Recent trends in microRNA research into breast cancer with particular focus on the associations between microRNAs and intrinsic subtypes. J Hum Genet. 2017;62:15–24.
Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14.
Itesako T, Seki N, Yoshino H, Chiyomaru T, Yamasaki T, Hidaka H, et al. The microRNA expression signature of bladder cancer by deep sequencing: the functional significance of the miR-195/497 cluster. PLoS ONE. 2014;9:e84311.
Goto Y, Kurozumi A, Arai T, Nohata N, Kojima S, Okato A, et al. Impact of novel miR-145-3p regulatory networks on survival in patients with castration-resistant prostate cancer. Br J Cancer. 2017;117:409–20.
Yonemori K, Seki N, Idichi T, Kurahara H, Osako Y, Koshizuka K, et al. The microRNA expression signature of pancreatic ductal adenocarcinoma by RNA sequencing: anti-tumour functions of the microRNA-216 cluster. Oncotarget. 2017;8:70097–115.
Koshizuka K, Nohata N, Hanazawa T, Kikkawa N, Arai T, Okato A, et al. Deep sequencing-based microRNA expression signatures in head and neck squamous cell carcinoma: dual strands of pre-miR-150 as antitumor miRNAs. Oncotarget. 2017;8:30288–304.
Arai T, Fuse M, Goto Y, Kaga K, Kurozumi A, Yamada Y, et al. Molecular pathogenesis of interstitial cystitis based on microRNA expression signature: miR-320 family-regulated molecular pathways and targets. J Hum Genet. 2018;63:543–54.
Idichi T, Seki N, Kurahara H, Yonemori K, Osako Y, Arai T, et al. Regulation of actin-binding protein ANLN by antitumor miR-217 inhibits cancer cell aggressiveness in pancreatic ductal adenocarcinoma. Oncotarget. 2017;8:53180–93.
Idichi T, Seki N, Kurahara H, Fukuhisa H, Toda H, Shimonosono M, et al. Molecular pathogenesis of pancreatic ductal adenocarcinoma: impact of passenger strand of pre-miR-148a on gene regulation. Cancer Sci. 2018;109:2013–26.
Idichi T, Seki N, Kurahara H, Fukuhisa H, Toda H, Shimonosono M, et al. Involvement of anti-tumor miR-124-3p and its targets in the pathogenesis of pancreatic ductal adenocarcinoma: direct regulation of ITGA3 and ITGB1 by miR-124-3p. Oncotarget. 2018;9:28849–65.
Yamada Y, Arai T, Sugawara S, Okato A, Kato M, Kojima S, et al. Impact of novel oncogenic pathways regulated by antitumor miR-451a in renal cell carcinoma. Cancer Sci. 2018;109:1239–53.
Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5–18.
Koshizuka K, Hanazawa T, Arai T, Okato A, Kikkawa N, Seki N. Involvement of aberrantly expressed microRNAs in the pathogenesis of head and neck squamous cell carcinoma. Cancer Metastas- Rev. 2017;36:525–45.
Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastas- Rev. 2013;32:623–42.
Goto Y, Kurozumi A, Nohata N, Kojima S, Matsushita R, Yoshino H, et al. The microRNA signature of patients with sunitinib failure: regulation of UHRF1 pathways by microRNA-101 in renal cell carcinoma. Oncotarget. 2016;7:59070–86.
Goto Y, Kojima S, Nishikawa R, Kurozumi A, Kato M, Enokida H, et al. MicroRNA expression signature of castration-resistant prostate cancer: the microRNA-221/222 cluster functions as a tumour suppressor and disease progression marker. Br J Cancer. 2015;113:1055–65.
Mizuno K, Mataki H, Arai T, Okato A, Kamikawaji K, Kumamoto T, et al. The microRNA expression signature of small cell lung cancer: tumor suppressors of miR-27a-5p and miR-34b-3p and their targeted oncogenes. J Hum Genet. 2017;62:671–8.
Peschansky VJ, Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics. 2014;9:3–12.
Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24:R762–776.
Arai T, Okato A, Yamada Y, Sugawara S, Kurozumi A, Kojima S, et al. Regulation of NCAPG by miR-99a-3p (passenger strand) inhibits cancer cell aggressiveness and is involved in CRPC. Cancer Med. 2018;7:1988–2002.
Chung TK, Lau TS, Cheung TH, Yim SF, Lo KW, Siu NS, et al. Dysregulation of microRNA-204 mediates migration and invasion of endometrial cancer by regulating FOXC1. Int J Cancer. 2012;130:1036–45.
Ding M, Lin B, Li T, Liu Y, Li Y, Zhou X, et al. A dual yet opposite growth-regulating function of miR-204 and its target XRN1 in prostate adenocarcinoma cells and neuroendocrine-like prostate cancer cells. Oncotarget. 2015;6:7686–7700.
Shi L, Zhang B, Sun X, Lu S, Liu Z, Liu Y, et al. MiR-204 inhibits human NSCLC metastasis through suppression of NUAK1. Br J Cancer. 2014;111:2316–27.
Yin Y, Zhang B, Wang W, Fei B, Quan C, Zhang J, et al. miR-204-5p inhibits proliferation and invasion and enhances chemotherapeutic sensitivity of colorectal cancer cells by downregulating RAB22A. Clin Cancer Res. 2014;20:6187–99.
Zeng J, Wei M, Shi R, Cai C, Liu X, Li T, et al. MiR-204-5p/Six1 feedback loop promotes epithelial−mesenchymal transition in breast cancer. Tumour Biol. 2016;37:2729–35.
Micalizzi DS, Christensen KL, Jedlicka P, Coletta RD, Baron AE, Harrell JC, et al. The Six1 homeoprotein induces human mammary carcinoma cells to undergo epithelial-mesenchymal transition and metastasis in mice through increasing TGF-beta signaling. J Clin Invest. 2009;119:2678–90.
Pliarchopoulou K, Kalogeras KT, Kronenwett R, Wirtz RM, Eleftheraki AG, Batistatou A, et al. Prognostic significance of RACGAP1 mRNA expression in high-risk early breast cancer: a study in primary tumors of breast cancer patients participating in a randomized Hellenic Cooperative Oncology Group trial. Cancer Chemother Pharmacol. 2013;71:245–55.
Milde-Langosch K, Karn T, Muller V, Witzel I, Rody A, Schmidt M, et al. Validity of the proliferation markers Ki67, TOP2A, and RacGAP1 in molecular subgroups of breast cancer. Breast Cancer Res Treat. 2013;137:57–67.
Kotoula V, Kalogeras KT, Kouvatseas G, Televantou D, Kronenwett R, Wirtz RM, et al. Sample parameters affecting the clinical relevance of RNA biomarkers in translational breast cancer research. Virchows Arch. 2013;462:141–54.
Glyvuk N, Tsytsyura Y, Geumann C, D'Hooge R, Huve J, Kratzke M, et al. AP-1/sigma1B-adaptin mediates endosomal synaptic vesicle recycling, learning and memory. EMBO J. 2010;29:1318–30.
Robinson MS, Bonifacino JS. Adaptor-related proteins. Curr Opin Cell Biol. 2001;13:444–53.
Ko SH, Jeon JI, Myung HS, Kim YJ, Kim JM. Bacteroides fragilis enterotoxin induces formation of autophagosomes in endothelial cells but interferes with fusion with lysosomes for complete autophagic flux through a mitogen-activated protein kinase-, AP-1-, and C/EBP homologous protein-dependent pathway. Infect Immun. 2017;85:e00420–17.
Setta-Kaffetzi N, Simpson MA, Navarini AA, Patel VM, Lu HC, Allen MH, et al. AP1S3 mutations are associated with pustular psoriasis and impaired Toll-like receptor 3 trafficking. Am J Hum Genet. 2014;94:790–7.
Mahil SK, Twelves S, Farkas K, Setta-Kaffetzi N, Burden AD, Gach JE, et al. AP1S3 mutations cause skin autoinflammation by disrupting keratinocyte autophagy and up-regulating IL-36 production. J Invest Dermatol. 2016;136:2251–9.
Frasor J, Chang EC, Komm B, Lin CY, Vega VB, Liu ET, et al. Gene expression preferentially regulated by tamoxifen in breast cancer cells and correlations with clinical outcome. Cancer Res. 2006;66:7334–40.
Li Y, Zou L, Li Q, Haibe-Kains B, Tian R, Li Y, Desmedt C, et al. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat Med. 2010;16:214–8.
Bergamaschi A, Christensen BL, Katzenellenbogen BS. Reversal of endocrine resistance in breast cancer: interrelationships among 14-3-3ζ, FOXM1, and a gene signature associated with mitosis. Breast Cancer Res. 2011;13:R70.
Wang W, Zhang L, Wang Y, Ding Y, Chen T, Wang Y, et al. Involvement of miR-451 in resistance to paclitaxel by regulating YWHAZ in breast cancer. Cell Death Dis. 2017;8:e3071.
Acknowledgements
The present study was supported by KAKENHI grants 15K10801, 18K09338, 16K19906, 17H04285, 16K10465 and 18K16322.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest
Rights and permissions
About this article
Cite this article
Toda, H., Kurozumi, S., Kijima, Y. et al. Molecular pathogenesis of triple-negative breast cancer based on microRNA expression signatures: antitumor miR-204-5p targets AP1S3. J Hum Genet 63, 1197–1210 (2018). https://doi.org/10.1038/s10038-018-0510-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-018-0510-3
This article is cited by
-
Plasma cell signatures predict prognosis and treatment efficacy for lung adenocarcinoma
Cellular Oncology (2024)
-
Endosomal protein expression of γ1-adaptin is associated with tumor growth activity and relapse-free survival in breast cancer
Breast Cancer (2024)
-
An integrated pan-cancer analysis of leucine-rich repeat containing protein 59: a potential biomarker for prognostic and immunotherapy
Genome Instability & Disease (2023)
-
Research on correlations of miR-585 expression with progression and prognosis of triple-negative breast cancer
Clinical and Experimental Medicine (2022)
-
MACE-Seq-based coding RNA and TrueQuant-based small RNA profile in breast cancer: tumor-suppressive miRNA-1275 identified as a novel marker
BMC Cancer (2021)