Abstract
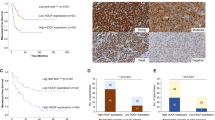
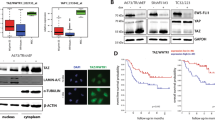
Ewing Sarcoma is the second most common solid pediatric malignant neoplasm of bone and soft tissue. Driven by EWS/Ets, or rarely variant, oncogenic fusions, Ewing Sarcoma is a biologically and clinically aggressive disease with a high propensity for metastasis. However, the mechanisms underpinning Ewing Sarcoma metastasis are currently not well understood. In the present study, we identify and characterize a novel metastasis-promotional pathway in Ewing Sarcoma, involving the histone demethylase KDM3A, previously identified by our laboratory as a new cancer-promoting gene in this disease. Using global gene expression profiling, we show that KDM3A positively regulates genes and pathways implicated in cell migration and metastasis, and demonstrate, using functional assays, that KDM3A promotes migration in vitro and experimental, post-intravasation, metastasis in vivo. We further identify the melanoma cell adhesion molecule (MCAM) as a novel KDM3A target gene in Ewing Sarcoma, and an important effector of KDM3A pro-metastatic action. Specifically, we demonstrate that MCAM depletion, like KDM3A depletion, inhibits cell migration in vitro and experimental metastasis in vivo, and that MCAM partially rescues impaired migration due to KDM3A knock-down. Mechanistically, we show that KDM3A regulates MCAM expression both through a direct mechanism, involving modulation of H3K9 methylation at the MCAM promoter, and an indirect mechanism, via the Ets1 transcription factor. Finally, we identify an association between high MCAM levels in patient tumors and poor survival, in two different Ewing Sarcoma clinical cohorts. Taken together, our studies uncover a new metastasis-promoting pathway in Ewing Sarcoma, with therapeutically targetable components.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Hospital SJCsR Disease Information, Solid Tumor: Ewing Sarcoma Family Tumors.
Gaspar N, Hawkins DS, Dirksen U, Lewis IJ, Ferrari S, Le Deley MC et al. Ewing sarcoma: current management and future approaches through collaboration. J Clin Oncol 2015; 33: 3036–3046.
Rodriguez-Galindo C, Navid F, Liu T, Billups CA, Rao BN, Krasin MJ . Prognostic factors for local and distant control in Ewing sarcoma family of tumors. Ann Oncol 2008; 19: 814–820.
Esiashvili N, Goodman M, Marcus RB Jr . Changes in incidence and survival of Ewing sarcoma patients over the past 3 decades: Surveillance Epidemiology and End Results data. J Pediatr Hematol Oncol 2008; 30: 425–430.
Grohar PJ, Helman LJ . Prospects and challenges for the development of new therapies for Ewing sarcoma. Pharmacol Ther 2013; 137: 216–224.
Choy E, Butrynski JE, Harmon DC, Morgan JA, George S, Wagner AJ et al. Phase II study of olaparib in patients with refractory Ewing sarcoma following failure of standard chemotherapy. BMC Cancer 2014; 14: 813.
Burchill SA . Ewing's sarcoma: diagnostic, prognostic, and therapeutic implications of molecular abnormalities. J Clin Pathol 2003; 56: 96–102.
Riggi N, Knoechel B, Gillespie SM, Rheinbay E, Boulay G, Suva ML et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell 2014; 26: 668–681.
Sankar S, Bell R, Stephens B, Zhuo R, Sharma S, Bearss DJ et al. Mechanism and relevance of EWS/FLI-mediated transcriptional repression in Ewing sarcoma. Oncogene 2013; 32: 5089–5100.
May WA, Lessnick SL, Braun BS, Klemsz M, Lewis BC, Lunsford LB et al. The Ewing's sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol Cell Biol 1993; 13: 7393–7398.
Prieur A, Tirode F, Cohen P, Delattre O . EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol Cell Biol 2004; 24: 7275–7283.
Chaturvedi A, Hoffman LM, Welm AL, Lessnick SL, Beckerle MC . The EWS/FLI oncogene drives changes in cellular morphology, adhesion, and migration in Ewing Sarcoma. Genes Cancer 2012; 3: 102–116.
Fadul J, Bell R, Hoffman LM, Beckerle MC, Engel ME, Lessnick SL . EWS/FLI utilizes NKX2-2 to repress mesenchymal features of Ewing sarcoma. Genes Cancer 2015; 6: 129–143.
Pedersen EA, Menon R, Bailey KM, Thomas DG, Van Noord RA, Tran J et al. Activation of Wnt/beta-catenin in ewing sarcoma cells antagonizes EWS/ETS function and promotes phenotypic transition to more metastatic cell states. Cancer Res 2016; 76: 5040–5053.
Feinberg AP, Koldobskiy MA, Gondor A . Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet 2016; 17: 284–299.
Lawlor ER, Thiele CJ . Epigenetic changes in pediatric solid tumors: promising new targets. Clin Cancer Res 2012; 18: 2768–2779.
Parrish JK, Sechler M, Winn RA, Jedlicka P . The histone demethylase KDM3A is a microRNA-22-regulated tumor promoter in Ewing Sarcoma. Oncogene 2015; 34: 257–262.
McFarlane S, Coulter JA, Tibbits P, O'Grady A, McFarlane C, Montgomery N et al. CD44 increases the efficiency of distant metastasis of breast cancer. Oncotarget 2015; 6: 11465–11476.
Gao Y, Ruan B, Liu W, Wang J, Yang X, Zhang Z et al. Knockdown of CD44 inhibits the invasion and metastasis of hepatocellular carcinoma both in vitro and in vivo by reversing epithelial-mesenchymal transition. Oncotarget 2015; 6: 7828–7837.
Wu GJ, Fu P, Wang SW, Wu MW . Enforced expression of MCAM/MUC18 increases in vitro motility and invasiveness and in vivo metastasis of two mouse melanoma K1735 sublines in a syngeneic mouse model. Mol Cancer Res 2008; 6: 1666–1677.
Xie S, Luca M, Huang S, Gutman M, Reich R, Johnson JP et al. Expression of MCAM/MUC18 by human melanoma cells leads to increased tumor growth and metastasis. Cancer Res 1997; 57: 2295–2303.
Tsai HC, Su HL, Huang CY, Fong YC, Hsu CJ, Tang CH . CTGF increases matrix metalloproteinases expression and subsequently promotes tumor metastasis in human osteosarcoma through down-regulating miR-519d. Oncotarget 2014; 5: 3800–3812.
Rappa G, Green TM, Karbanova J, Corbeil D, Lorico A . Tetraspanin CD9 determines invasiveness and tumorigenicity of human breast cancer cells. Oncotarget 2015; 6: 7970–7991.
Herr MJ, Kotha J, Hagedorn N, Smith B, Jennings LK . Tetraspanin CD9 promotes the invasive phenotype of human fibrosarcoma cells via upregulation of matrix metalloproteinase-9. PLoS One 2013; 8: e67766.
Miller NL, Lawson C, Chen XL, Lim ST, Schlaepfer DD . Rgnef (p190RhoGEF) knockout inhibits RhoA activity, focal adhesion establishment, and cell motility downstream of integrins. PLoS One 2012; 7: e37830.
Yu HG, Nam JO, Miller NL, Tanjoni I, Walsh C, Shi L et al. p190RhoGEF (Rgnef) promotes colon carcinoma tumor progression via interaction with focal adhesion kinase. Cancer Res 2011; 71: 360–370.
Pavon MA, Arroyo-Solera I, Tellez-Gabriel M, Leon X, Viros D, Lopez M et al. Enhanced cell migration and apoptosis resistance may underlie the association between high SERPINE1 expression and poor outcome in head and neck carcinoma patients. Oncotarget 2015; 6: 29016–29033.
Yu XM, Jaskula-Sztul R, Georgen MR, Aburjania Z, Somnay YR, Leverson G et al. Notch1 signaling regulates the aggressiveness of differentiated thyroid cancer and inhibits SERPINE1 expression. Clin Cancer Res 2016; 22: 3582–3592.
Sang Y, Chen MY, Luo D, Zhang RH, Wang L, Li M et al. TEL2 suppresses metastasis by down-regulating SERPINE1 in nasopharyngeal carcinoma. Oncotarget 2015; 6: 29240–29253.
Du C, Gao Y, Xu S, Jia J, Huang Z, Fan J et al. KLF5 promotes cell migration by up-regulating FYN in bladder cancer cells. FEBS Lett 2016; 590: 408–418.
Jia L, Zhou Z, Liang H, Wu J, Shi P, Li F et al. KLF5 promotes breast cancer proliferation, migration and invasion in part by upregulating the transcription of TNFAIP2. Oncogene 2016; 35: 2040–2051.
Li L, Zhang Z, Ma T, Huo R . PRMT1 regulates tumor growth and metastasis of human melanoma via targeting ALCAM. Mol Med Rep 2016; 14: 521–528.
Hansen AG, Arnold SA, Jiang M, Palmer TD, Ketova T, Merkel A et al. ALCAM/CD166 is a TGF-beta-responsive marker and functional regulator of prostate cancer metastasis to bone. Cancer Res 2014; 74: 1404–1415.
Penna E, Orso F, Cimino D, Vercellino I, Grassi E, Quaglino E et al. miR-214 coordinates melanoma progression by upregulating ALCAM through TFAP2 and miR-148b downmodulation. Cancer Res 2013; 73: 4098–4111.
Shi W, Zhang C, Chen Z, Chen H, Liu L, Meng Z . Cyr61-positive cancer stem-like cells enhances distal metastases of pancreatic cancer. Oncotarget 2016; 7: 73160–73170.
Hou CH, Lin FL, Hou SM, Liu JF . Cyr61 promotes epithelial-mesenchymal transition and tumor metastasis of osteosarcoma by Raf-1/MEK/ERK/Elk-1/TWIST-1 signaling pathway. Mol Cancer 2014; 13: 236.
Habel N, Vilalta M, Bawa O, Opolon P, Blanco J, Fromigue O . Cyr61 silencing reduces vascularization and dissemination of osteosarcoma tumors. Oncogene 2015; 34: 3207–3213.
Oskarsson T, Acharyya S, Zhang XH, Vanharanta S, Tavazoie SF, Morris PG et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med 2011; 17: 867–874.
Paron I, Berchtold S, Voros J, Shamarla M, Erkan M, Hofler H et al. Tenascin-C enhances pancreatic cancer cell growth and motility and affects cell adhesion through activation of the integrin pathway. PLoS One 2011; 6: e21684.
Dittmer J . The role of the transcription factor Ets1 in carcinoma. Semin Cancer Biol 2015; 35: 20–38.
Zheng L, Qi T, Yang D, Qi M, Li D, Xiang X et al. microRNA-9 suppresses the proliferation, invasion and metastasis of gastric cancer cells through targeting cyclin D1 and Ets1. PLoS One 2013; 8: e55719.
Lim SY, Yuzhalin AE, Gordon-Weeks AN, Muschel RJ . Targeting the CCL2-CCR2 signaling axis in cancer metastasis. Oncotarget 2016; 7: 28697–28710.
Yang J, Lv X, Chen J, Xie C, Xia W, Jiang C et al. CCL2-CCR2 axis promotes metastasis of nasopharyngeal carcinoma by activating ERK1/2-MMP2/9 pathway. Oncotarget 2016; 7: 15632–15647.
Kitamura T, Qian BZ, Soong D, Cassetta L, Noy R, Sugano G et al. CCL2-induced chemokine cascade promotes breast cancer metastasis by enhancing retention of metastasis-associated macrophages. J Exp Med 2015; 212: 1043–1059.
Park JS, Lee JH, Lee YS, Kim JK, Dong SM, Yoon DS . Emerging role of LOXL2 in the promotion of pancreas cancer metastasis. Oncotarget 2016; 7: 42539–42552.
Peng DH, Ungewiss C, Tong P, Byers LA, Wang J, Canales JR et al. ZEB1 induces LOXL2-mediated collagen stabilization and deposition in the extracellular matrix to drive lung cancer invasion and metastasis. Oncogene 2016, e-pub ahead of print 3 October 2016.
Moreno-Bueno G, Salvador F, Martin A, Floristan A, Cuevas EP, Santos V et al. Lysyl oxidase-like 2 (LOXL2), a new regulator of cell polarity required for metastatic dissemination of basal-like breast carcinomas. EMBO Mol Med 2011; 3: 528–544.
Liang XH, Zhang GX, Zeng YB, Yang HF, Li WH, Liu QL et al. LIM protein JUB promotes epithelial-mesenchymal transition in colorectal cancer. Cancer Sci 2014; 105: 660–666.
Hou Z, Peng H, Ayyanathan K, Yan KP, Langer EM, Longmore GD et al. The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol 2008; 28: 3198–3207.
Wang Z, Yan X . CD146, a multi-functional molecule beyond adhesion. Cancer Lett 2013; 330: 150–162.
Orentas RJ, Yang JJ, Wen X, Wei JS, Mackall CL, Khan J . Identification of cell surface proteins as potential immunotherapy targets in 12 pediatric cancers. Front Oncol 2012; 2: 194.
Volchenboum SL, Andrade J, Huang L, Barkauskas DA, Krailo M, Womer RB et al. Gene expression profiling of ewing sarcoma tumors reveals the prognostic importance of tumor-stromal interactions: a report from the children's oncology group. J Pathol Clin Res 2015; 1: 83–94.
Douglas D, Hsu JH, Hung L, Cooper A, Abdueva D, van Doorninck J et al. BMI-1 promotes ewing sarcoma tumorigenicity independent of CDKN2A repression. Cancer Res 2008; 68: 6507–6515.
Richter GH, Plehm S, Fasan A, Rossler S, Unland R, Bennani-Baiti IM et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc Natl Acad Sci USA 2009; 106: 5324–5329.
Bennani-Baiti IM, Machado I, Llombart-Bosch A, Kovar H . Lysine-specific demethylase 1 (LSD1/KDM1A/AOF2/BHC110) is expressed and is an epigenetic drug target in chondrosarcoma, Ewing's sarcoma, osteosarcoma, and rhabdomyosarcoma. Hum Pathol 2012; 43: 1300–1307.
Patel M, Simon JM, Iglesia MD, Wu SB, McFadden AW, Lieb JD et al. Tumor-specific retargeting of an oncogenic transcription factor chimera results in dysregulation of chromatin and transcription. Genome Res 2012; 22: 259–270.
Svoboda LK, Harris A, Bailey NJ, Schwentner R, Tomazou E, von Levetzow C et al. Overexpression of HOX genes is prevalent in Ewing sarcoma and is associated with altered epigenetic regulation of developmental transcription programs. Epigenetics 2014; 9: 1613–1625.
Ramadoss S, Sen S, Ramachandran I, Roy S, Chaudhuri G, Farias-Eisner R . Lysine-specific demethylase KDM3A regulates ovarian cancer stemness and chemoresistance. Oncogene 2016, e-pub ahead of print 3 October 2016.
Ramadoss S, Guo G, Wang CY . Lysine demethylase KDM3A regulates breast cancer cell invasion and apoptosis by targeting histone and the non-histone protein p53. Oncogene 2016; 36: 47–59.
Mahajan K, Lawrence HR, Lawrence NJ, Mahajan NP . ACK1 tyrosine kinase interacts with histone demethylase KDM3A to regulate the mammary tumor oncogene HOXA1. J Biol Chem 2014; 289: 28179–28191.
Cho HS, Toyokawa G, Daigo Y, Hayami S, Masuda K, Ikawa N et al. The JmjC domain-containing histone demethylase KDM3A is a positive regulator of the G1/S transition in cancer cells via transcriptional regulation of the HOXA1 gene. Int J Cancer 2012; 131: E179–E189.
Tee AE, Ling D, Nelson C, Atmadibrata B, Dinger ME, Xu N et al. The histone demethylase JMJD1A induces cell migration and invasion by up-regulating the expression of the long noncoding RNA MALAT1. Oncotarget 2014; 5: 1793–1804.
Ohguchi H, Hideshima T, Bhasin MK, Gorgun GT, Santo L, Cea M et al. The KDM3A-KLF2-IRF4 axis maintains myeloma cell survival. Nat Commun 2016; 7: 10258.
Shih IM, Elder DE, Speicher D, Johnson JP, Herlyn M . Isolation and functional characterization of the A32 melanoma-associated antigen. Cancer Res 1994; 54: 2514–2520.
Lin Y, Wu X, Shen Y, Bu P, Yang D, Yan X . A novel antibody AA98 V(H)/L directed against CD146 efficiently inhibits angiogenesis. Anticancer Res 2007; 27: 4219–4224.
Jouve N, Despoix N, Espeli M, Gauthier L, Cypowyj S, Fallague K et al. The involvement of CD146 and its novel ligand Galectin-1 in apoptotic regulation of endothelial cells. J Biol Chem 2013; 288: 2571–2579.
Bardin N, Blot-Chabaud M, Despoix N, Kebir A, Harhouri K, Arsanto JP et al. CD146 and its soluble form regulate monocyte transendothelial migration. Arterioscler Thromb Vasc Biol 2009; 29: 746–753.
Zhang X, Wang Z, Kang Y, Li X, Ma X, Ma L . MCAM expression is associated with poor prognosis in non-small cell lung cancer. Clin Transl Oncol 2014; 16: 178–183.
Zeng Q, Li W, Lu D, Wu Z, Duan H, Luo Y et al. CD146, an epithelial-mesenchymal transition inducer, is associated with triple-negative breast cancer. Proc Natl Acad Sci USA 2012; 109: 1127–1132.
Imbert AM, Garulli C, Choquet E, Koubi M, Aurrand-Lions M, Chabannon C . CD146 expression in human breast cancer cell lines induces phenotypic and functional changes observed in Epithelial to Mesenchymal Transition. PLoS One 2012; 7: e43752.
Jouve N, Bachelier R, Despoix N, Blin MG, Matinzadeh MK, Poitevin S et al. CD146 mediates VEGF-induced melanoma cell extravasation through FAK activation. Int J Cancer 2015; 137: 50–60.
Liu XS, Genet MD, Haines JE, Mehanna EK, Wu S, Chen HI et al. zbtb7a suppresses melanoma metastasis by transcriptionally repressing MCAM. Mol Cancer Res 2015; 13: 1206–1217.
Schiano C, Grimaldi V, Casamassimi A, Infante T, Esposito A, Giovane A et al. Different expression of CD146 in human normal and osteosarcoma cell lines. Med Oncol 2012; 29: 2998–3002.
Dittmer J . The biology of the Ets1 proto-oncogene. Mol Cancer 2003; 2: 29.
Heidenreich B, Rachakonda PS, Hemminki K, Kumar R . TERT promoter mutations in cancer development. Curr Opin Genet Dev 2014; 24: 30–37.
Zhan M, Wen F, Liu L, Chen Z, Wei H, Zhou H . JMJD1A promotes tumorigenesis and forms a feedback loop with EZH2/let-7c in NSCLC cells. Tumour Biol 2016; 37: 11237–11247.
Yang H, Liu Z, Yuan C, Zhao Y, Wang L, Hu J et al. Elevated JMJD1A is a novel predictor for prognosis and a potential therapeutic target for gastric cancer. Int J Clin Exp Pathol 2015; 8: 11092–11099.
Wade MA, Jones D, Wilson L, Stockley J, Coffey K, Robson CN et al. The histone demethylase enzyme KDM3A is a key estrogen receptor regulator in breast cancer. Nucleic Acids Res 2015; 43: 196–207.
Pa M, Naizaer G, Seyiti A, Kuerbang G . KDM3A confers metastasis and chemoresistance in epithelial ovarian cancer. J Mol Histol 2015; 47: 103.
Sohni A, Verfaillie CM . Mesenchymal stem cells migration homing and tracking. Stem Cells Int 2013; 2013: 130763.
Steingen C, Brenig F, Baumgartner L, Schmidt J, Schmidt A, Bloch W . Characterization of key mechanisms in transmigration and invasion of mesenchymal stem cells. J Mol Cell Cardiol 2008; 44: 1072–1084.
Smith R, Owen LA, Trem DJ, Wong JS, Whangbo JS, Golub TR et al. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing's sarcoma. Cancer Cell 2006; 9: 405–416.
Chaturvedi A, Hoffman LM, Jensen CC, Lin YC, Grossmann AH, Randall RL et al. Molecular dissection of the mechanism by which EWS/FLI expression compromises actin cytoskeletal integrity and cell adhesion in Ewing sarcoma. Mol Biol Cell 2014; 25: 2695–2709.
Owen LA, Kowalewski AA, Lessnick SL . EWS/FLI mediates transcriptional repression via NKX2.2 during oncogenic transformation in Ewing's sarcoma. PLoS One 2008; 3: e1965.
Li X, Xu Z, Du W, Zhang Z, Wei Y, Wang H et al. Aiolos promotes anchorage independence by silencing p66Shc transcription in cancer cells. Cancer Cell 2014; 25: 575–589.
Wu ZJ, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F . A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc 2004; 99: 909–917.
Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 2003; 4: P3.
Huang, da W, Sherman BT, Lempicki RA . Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25: 25–29.
McKinsey EL, Parrish JK, Irwin AE, Niemeyer BF, Kern HB, Birks DK et al. A novel oncogenic mechanism in Ewing sarcoma involving IGF pathway targeting by EWS/Fli1-regulated microRNAs. Oncogene 2011; 30: 4910–4920.
Sarbassov DD, Guertin DA, Ali SM, Sabatini DM . Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 2005; 307: 1098–1101.
Ponomarev V, Doubrovin M, Serganova I, Vider J, Shavrin A, Beresten T et al. A novel triple-modality reporter gene for whole-body fluorescent, bioluminescent, and nuclear noninvasive imaging. Eur J Nucl Med Mol Imaging 2004; 31: 740–751.
Acknowledgements
We wish to thank Elizabeth Wellberg for assistance with gene expression profiling studies; members of the Tobias Neff and Kathrin Bernt laboratories for assistance with ChIP studies; Elizabeth Lawlor at the University of Michigan for the TC32 cell line, and tumor expression profiling and patient outcome data; and Steve Lessnick at Nationwide Children’s Hospital for retroviral packaging constructs. We further wish to thank the University of Colorado Cancer Center Flow Cytometry, Functional Genomics, Tissue Culture, DNA Sequencing and Small Animal Imaging Core Facilities, supported by P30-CA046934. Funding support for this work was provided by the Front Range Cancer Challenge, Children’s Hospital Colorado Research Institute, University of Colorado School of Medicine Academic Enrichment Funds, and R01-CA183874 (PJ); the Cancer League of Colorado (PJ and MS); and F31-CA203053 (MS).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Sechler, M., Parrish, J., Birks, D. et al. The histone demethylase KDM3A, and its downstream target MCAM, promote Ewing Sarcoma cell migration and metastasis. Oncogene 36, 4150–4160 (2017). https://doi.org/10.1038/onc.2017.44
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.44
This article is cited by
-
Knockout of KDM3A in MDA-MB-231 breast cancer cells inhibits tumor malignancy and promotes apoptosis
Journal of Molecular Histology (2024)
-
Myeloid cells interact with a subset of thyrocytes to promote their migration and follicle formation through NF-κB
Nature Communications (2023)
-
The proteomic landscape of soft tissue sarcomas
Nature Communications (2023)
-
KDM3A-mediated SP1 activates PFKFB4 transcription to promote aerobic glycolysis in osteosarcoma and augment tumor development
BMC Cancer (2022)
-
The histone demthylase KDM3A protects the myocardium from ischemia/reperfusion injury via promotion of ETS1 expression
Communications Biology (2022)