Abstract
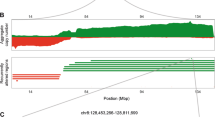
Chromosomal translocation is the best-characterized genetic mechanism for oncogene activation. However, there are documented examples of activation by alternate mechanisms, for example gene dosage increase, though its prevalence is unclear. Here, we answered the fundamental question of the contribution of DNA amplification as a molecular mechanism driving oncogenesis. Comparing 104 cancer lines representing diverse tissue origins identified genes residing in amplification ‘hotspots’ and discovered an unexpected frequency of genes activated by this mechanism. The 3431 amplicons identified represent ∼10 per hematological and ∼36 per epithelial cancer genome. Many recurrently amplified oncogenes were previously known to be activated only by disease-specific translocations. The 135 hotspots identified contain 538 unique genes and are enriched for proliferation, apoptosis and linage-dependency genes, reflecting functions advantageous to tumor growth. Integrating gene dosage with expression data validated the downstream impact of the novel amplification events in both cell lines and clinical samples. For example, multiple downstream components of the EGFR-family-signaling pathway, including CDK5, AKT1 and SHC1, are overexpressed as a direct result of gene amplification in lung cancer. Our findings suggest that amplification is far more common a mechanism of oncogene activation than previously believed and that specific regions of the genome are hotspots of amplification.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Albertson DG . (2006). Gene amplification in cancer. Trends Genet 22: 447–455.
Apergis GA, Crawford N, Ghosh D, Steppan CM, Vorachek WR, Wen P et al. (1998). A novel nk-2-related transcription factor associated with human fetal liver and hepatocellular carcinoma. J Biol Chem 273: 2917–2925.
Boersma-Vreugdenhil GR, Kuipers J, Van Stralen E, Peeters T, Michaux L, Hagemeijer A et al. (2004). The recurrent translocation t(14;20)(q32;q12) in multiple myeloma results in aberrant expression of MAFB: a molecular and genetic analysis of the chromosomal breakpoint. Br J Haematol 126: 355–363.
Bublil EM, Yarden Y . (2007). The EGF receptor family: spearheading a merger of signaling and therapeutics. Curr Opin Cell Biol 19: 124–134.
Buttel I, Fechter A, Schwab M . (2004). Common fragile sites and cancer: targeted cloning by insertional mutagenesis. Ann NY Acad Sci 1028: 14–27.
Chari R, Lockwood WW, Coe BP, Chu A, Macey D, Thomson A et al. (2006). SIGMA: a system for integrative genomic microarray analysis of cancer genomes. BMC Genomics 7: 324.
Chi B, DeLeeuw RJ, Coe BP, MacAulay C, Lam WL . (2004). SeeGH—a software tool for visualization of whole genome array comparative genomic hybridization data. BMC Bioinformatics 5: 13.
Coe BP, Lockwood WW, Girard L, Chari R, Macaulay C, Lam S et al. (2006). Differential disruption of cell cycle pathways in small cell and non-small cell lung cancer. Br J Cancer 94: 1927–1935.
Coe BP, Ylstra B, Carvalho B, Meijer GA, Macaulay C, Lam WL . (2007). Resolving the resolution of array CGH. Genomics 89: 647–653.
Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO et al. (2007). MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316: 1039–1043.
Fabbro D, Di Loreto C, Stamerra O, Beltrami CA, Lonigro R, Damante G . (1996). TTF-1 gene expression in human lung tumours. Eur J Cancer 32A: 512–517.
Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R et al. (2004). A census of human cancer genes. Nat Rev Cancer 4: 177–183.
Garnis C, Lockwood WW, Vucic E, Ge Y, Girard L, Minna JD et al. (2006). High resolution analysis of non-small cell lung cancer cell lines by whole genome tiling path array CGH. Int J Cancer 118: 1556–1564.
Garraway LA, Sellers WR . (2006). Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer 6: 593–602.
Greshock J, Nathanson K, Martin AM, Zhang L, Coukos G, Weber BL et al. (2007). Cancer cell lines as genetic models of their parent histology: analyses based on array comparative genomic hybridization. Cancer Res 67: 3594–3600.
Heidenblad M, Lindgren D, Veltman JA, Jonson T, Mahlamaki EH, Gorunova L et al. (2005). Microarray analyses reveal strong influence of DNA copy number alterations on the transcriptional patterns in pancreatic cancer: implications for the interpretation of genomic amplifications. Oncogene 24: 1794–1801.
Hellman A, Zlotorynski E, Scherer SW, Cheung J, Vincent JB, Smith DI et al. (2002). A role for common fragile site induction in amplification of human oncogenes. Cancer Cell 1: 89–97.
Hemstrom TH, Sandstrom M, Zhivotovsky B . (2006). Inhibitors of the PI3-kinase/Akt pathway induce mitotic catastrophe in non-small cell lung cancer cells. Int J Cancer 119: 1028–1038.
Henderson LJ, Coe BP, Lee EH, Girard L, Gazdar AF, Minna JD et al. (2005). Genomic and gene expression profiling of minute alterations of chromosome arm 1p in small-cell lung carcinoma cells. Br J Cancer 92: 1553–1560.
Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB . (2005). Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov 4: 988–1004.
Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E et al. (2002). Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res 62: 6240–6245.
Ishkanian AS, Malloff CA, Watson SK, DeLeeuw RJ, Chi B, Coe BP et al. (2004). A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet 36: 299–303.
Khojasteh M, Lam WL, Ward RK, MacAulay C . (2005). A stepwise framework for the normalization of array CGH data. BMC Bioinformatics 6: 274.
Koizumi F, Shimoyama T, Taguchi F, Saijo N, Nishio K . (2005). Establishment of a human non-small cell lung cancer cell line resistant to gefitinib. Int J Cancer 116: 36–44.
Lockwood WW, Coe BP, Williams AC, MacAulay C, Lam WL . (2007). Whole genome tiling path array CGH analysis of segmental copy number alterations in cervical cancer cell lines. Int J Cancer 120: 436–443.
Mitelman F . (2000). Recurrent chromosome aberrations in cancer. Mutat Res 462: 247–253.
Myllykangas S, Himberg J, Bohling T, Nagy B, Hollmen J, Knuutila S . (2006). DNA copy number amplification profiling of human neoplasms. Oncogene 25: 7324–7332.
Myllykangas S, Knuutila S . (2006). Manifestation, mechanisms and mysteries of gene amplifications. Cancer Lett 232: 79–89.
Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T et al. (2006). A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10: 515–527.
Pierotti MA, Bongarzone I, Borello MG, Greco A, Pilotti S, Sozzi G . (1996). Cytogenetics and molecular genetics of carcinomas arising from thyroid epithelial follicular cells. Genes Chromosomes Cancer 16: 1–14.
Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE et al. (2002). Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci USA 99: 12963–12968.
Roccato E, Bressan P, Sabatella G, Rumio C, Vizzotto L, Pierotti MA et al. (2005). Proximity of TPR and NTRK1 rearranging loci in human thyrocytes. Cancer Res 65: 2572–2576.
Squire JA, Pei J, Marrano P, Beheshti B, Bayani J, Lim G et al. (2003). High-resolution mapping of amplifications and deletions in pediatric osteosarcoma by use of CGH analysis of cDNA microarrays. Genes Chromosomes Cancer 38: 215–225.
Stenhouse G, Fyfe N, King G, Chapman A, Kerr KM . (2004). Thyroid transcription factor 1 in pulmonary adenocarcinoma. J Clin Pathol 57: 383–387.
Tanaka H, Yanagisawa K, Shinjo K, Taguchi A, Maeno K, Tomida S et al. (2007). Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res 67: 6007–6011.
Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y et al. (2005). High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci USA 102: 9625–9630.
Wahl GM, Robert de Saint Vincent B, DeRose ML . (1984). Effect of chromosomal position on amplification of transfected genes in animal cells. Nature 307: 516–520.
Wang PW, Eisenbart JD, Cordes SP, Barsh GS, Stoffel M, Le Beau MM . (1999). Human KRML (MAFB): cDNA cloning, genomic structure, and evaluation as a candidate tumor suppressor gene in myeloid leukemias. Genomics 59: 275–281.
Watson SK, deLeeuw RJ, Horsman DE, Squire JA, Lam WL . (2007). Cytogenetically balanced translocations are associated with focal copy number alterations. Hum Genet 120: 795–805.
Watson SK, deLeeuw RJ, Ishkanian AS, Malloff CA, Lam WL . (2004). Methods for high throughput validation of amplified fragment pools of BAC DNA for constructing high resolution CGH arrays. BMC Genomics 5: 6.
Weinstein IB . (2002). Cancer. Addiction to oncogenes—the Achilles heal of cancer. Science 297: 63–64.
Wolf M, Mousses S, Hautaniemi S, Karhu R, Huusko P, Allinen M et al. (2004). High-resolution analysis of gene copy number alterations in human prostate cancer using CGH on cDNA microarrays: impact of copy number on gene expression. Neoplasia 6: 240–247.
Zhou BB, Peyton M, He B, Liu C, Girard L, Caudler E et al. (2006). Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell 10: 39–50.
Acknowledgements
This work was supported by funds from CIHR, Genome Canada/BC, Lung Cancer SPORE P50CA70907, DOD VITAL, the Gillson Longenbaugh and Anderson Charitable Foundations as well as scholarships from NSERC, CIHR and MSFHR to WWL, RC and BPC.
Author information
Authors and Affiliations
Corresponding author
Additional information
Data deposition: Gene Expression Omnibus, accession number GSE-4824.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
Supplementary information
Rights and permissions
About this article
Cite this article
Lockwood, W., Chari, R., Coe, B. et al. DNA amplification is a ubiquitous mechanism of oncogene activation in lung and other cancers. Oncogene 27, 4615–4624 (2008). https://doi.org/10.1038/onc.2008.98
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2008.98
Keywords
This article is cited by
-
Assessment of MYC and TERT copy number variations in lung cancer using digital PCR
BMC Research Notes (2023)
-
Interpreting pathways to discover cancer driver genes with Moonlight
Nature Communications (2020)
-
The oncogenic neurotrophin receptor tropomyosin-related kinase variant, TrkAIII
Journal of Experimental & Clinical Cancer Research (2018)
-
Contrasting origin of B chromosomes in two cervids (Siberian roe deer and grey brocket deer) unravelled by chromosome-specific DNA sequencing
BMC Genomics (2016)
-
CDK5 functions as a tumor promoter in human colorectal cancer via modulating the ERK5–AP-1 axis
Cell Death & Disease (2016)