Abstract
We have previously demonstrated that the thiazole derivative 3-methylcyclopentylidene-[4-(4′-chlorophenyl)thiazol-2-yl]hydrazone (CPTH6) induces apoptosis and cell cycle arrest in human leukemia cells. The aim of this study was to evaluate whether CPTH6 is able to affect autophagy. By using several human tumor cell lines with different origins we demonstrated that CPTH6 treatment induced, in a dose-dependent manner, a significant increase in autophagic features, as imaged by electron microscopy, immunoblotting analysis of membrane-bound form of microtubule-associated protein 1 light chain 3 (LC3B-II) levels and by appearance of typical LC3B-II-associated autophagosomal puncta. To gain insights into the molecular mechanisms of elevated markers of autophagy induced by CPTH6 treatment, we silenced the expression of several proteins acting at different steps of autophagy. We found that the effect of CPTH6 on autophagy developed through a noncanonical mechanism that did not require beclin-1-dependent nucleation, but involved Atg-7-mediated elongation of autophagosomal membranes. Strikingly, a combined treatment of CPTH6 with late-stage autophagy inhibitors, such as chloroquine and bafilomycin A1, demonstrates that under basal condition CPTH6 reduces autophagosome turnover through an impairment of their degradation pathway, rather than enhancing autophagosome formation, as confirmed by immunofluorescence experiments. According to these results, CPTH6-induced enhancement of autophagy substrate p62 and NBR1 protein levels confirms a blockage of autophagic cargo degradation. In addition, CPTH6 inhibited autophagosome maturation and compounds having high structural similarities with CPTH6 produced similar effects on the autophagic pathway. Finally, the evidence that CPTH6 treatment decreased α-tubulin acetylation and failed to increase autophagic markers in cells in which acetyltransferase ATAT1 expression was silenced indicates a possible role of α-tubulin acetylation in CPTH6-induced alteration in autophagy. Overall, CPTH6 could be a valuable agent for the treatment of cancer and should be further studied as a possible antineoplastic agent.
Similar content being viewed by others
Main
Autophagy has been predominantly characterized as a nonselective degradative pathway activated by starvation. In this context, it is a process known to provide metabolic sources to the cell under nutrient depletion or other stresses. However, it has become apparent that autophagy is not solely dedicated to nutrient management, and a nutrient-independent ‘basal’ autophagy has been defined.1 It occurs at basal levels in most tissues and contributes to the routine turnover of cytoplasmic components, thus enforcing intracellular quality control.2
Induction of autophagy may be a double-edged sword, as it is also a mechanism leading to cell death when destruction of cell cytoplasm components is activated in a long-lasting way.3
Autophagy is triggered by various stimuli, is mediated by a number of proteins encoded by autophagy-related genes (Atgs) and is usually described as a cascade of events such as nucleation, elongation, maturation and fusion. A canonical mechanism of nucleation (de novo formation) of autophagosomal membranes depends on beclin-1, which interacts with adaptor protein p150 to stimulate activity of PI3-kinase VPS34.4 The elongation requires Atg-12, Atg-3, Atg-5 and Atg-7,5 whereas lysosome-associated membrane proteins 1 and 2 (LAMP-1 and LAMP-2) regulate the late step of the autophagic process.
Autophagic pathway can be inhibited by pharmacological inhibitors at different steps: PI3-kinase inhibitors block autophagosome formation;6 microtubule-disrupting agents and endoplasmic reticulum stressors inhibit autophagosome–lysosome fusion;7, 8 and lysosomal proteases inhibitors and acidification modulators strongly reduce final degradation of autophagic cargo inside autolysosome.9
Recently, the cross-talk between autophagy and apoptosis has been considered as a key factor in the development and treatment of cancer.3 The two pathways share molecular regulators and, in some cases, are activated by the same stimulus. Despite the great deal of interest in the regulation of autophagy for therapeutic purposes, there are only few modulators of the autophagic pathway that have shown promising pharmacological value.10, 11, 12
Recently, CPTH6 (3-methylcyclopentylidene-[4-(4′-chlorophenyl)thiazol-2-yl]hydrazone), a newly synthesized molecule derived from thiazole, has been characterized for its ability to activate apoptotic program in human acute myeloid leukemia cell lines (AML).13, 14 Here, by using either pharmacological or genetic means at the early or late stages of autophagy, we analyzed the effect of CPTH6 on autophagic pathway on a panel of human cancer cell lines.
Results
CPTH6 induces a block of basal autophagy
We previously demonstrated that tumor cell lines undergo apoptosis after CPTH6 treatment.14 Because many lines of evidence suggest a link between apoptosis and autophagy,15 in this paper we examined the effect of CPTH6 on autophagy in several tumor cell lines with different histotypes.
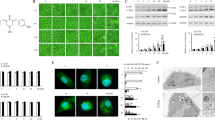
We first analyzed CPTH6-induced changes in the levels of autophagosomal marker microtubule-associated protein 1 light chain 3 (LC3B) in leukemia, melanoma, ovary and lung carcinoma cell lines, exposed to increasing concentrations of CPTH6 for 72 h (Figure 1A). Upon treatment with CPTH6, a significant increase in the amount of LC3B-II in a dose-dependent manner was observed, although to a different extent, in all cell lines.
CPTH6 treatment induces autophagic markers under basal conditions. (A) Western blot analysis of LC3B-I to LC3B-II conversion in the indicated cell lines after 72 h of treatment with CPTH6. HSP72/73 is shown as a loading and transferring control. Western blots representative of three independent experiments with similar results are shown. LC3B-II levels were quantified by densitometric analyses and fold increase relative to untreated cells are presented. (B) Representative images of fluorescence microscopy and (C) quantification of cells positive for autophagosomal structures in H1299 cells stably expressing EGFP-LC3B protein untreated (black) or treated with CPTH6 (100 μM, white) for the indicated times. The results represent the average±S.E.M. of three independent experiments. *P values were calculated between untreated and treated cells, P<0.05. (D) TEM analysis in H1299 cells (a) untreated and (b–d) treated with CPTH6 (100 μM, 24 h). Untreated cells showed their typical ultrastructural features with well-preserved cytoplasm, indented nucleus with prominent nucleolus and dispersed chromatin. After CPTH6 treatment, numerous vesicles containing material, including organelles, in various degrees of degradation are observed in the cytoplasm (b, see arrows). At higher magnification (c), myelin-like structures and not well-delimited autophagosomes induced by CPTH6 treatment were visible (box in the picture). Numerous lipid highly osmiophilic granules (see arrows) were also very often observed in the treated cells (d). N, nucleus
Formation of punctate spots (puncta) with enhanced green fluorescence protein (EGFP)-LC3B reporter is a well-characterized marker to visualize autophagosomes and represents the accumulation of LC3B-II on autophagic vesicles. Thus, we analyzed autophagosome formation in H1299 cells stably expressing EGFP-LC3B protein treated with 100 μM CPTH6 from 6 to 72 h. As shown in Figures 1B and C, a diffuse cytoplasmic distribution of green fluorescence was observed in untreated cells, whereas a characteristic redistribution of EGFP-LC3B from a diffused staining to puncta was evident, already 6 h after treatment, in a time-dependent manner. A dose-dependent effect was also observed in M14 cells (Supplementary Figure 1A, B).
The formation of autophagosomes induced in H1299 cells by CPTH6 treatment was also examined with transmission electron microscopy (TEM). As reported in Figure 1D, the induction of autophagy was witnessed by vacuolization of the cytoplasm because of cytotoxic treatment, not observed in the control cells. Only few and immature autophagosomes, characterized by an electron density equivalent to the cytoplasm, coexisting with late vesicles (primary and secondary lysosomes) were observed after 24 h of CPTH6 treatment. Treated cells did not contain double-membrane autophagic vacuoles, and the membrane structures observed in the cytoplasm may be attempting to form phagophores, which should have led to the construction of autophagosomes.
The increase in LC3B-II levels induced by CPTH6 treatment could be related to either enhanced autophagosome formation, due to an increase in autophagic activity, or reduced turnover of autophagosomes, due to an impairment of the degradation pathway.16, 17, 18 Only if autophagy is activated by CPTH6, late-stage autophagy inhibitors will be expected to further increase LC3B-II level and the number of autophagosomes. Therefore, to discriminate between the two possibilities, we utilized the early-stage autophagy inhibitor 3-methyladenine (3-MA) and the two late-stage inhibitors bafilomycin A1 and chloroquine. Autophagy inhibition by 3-MA completely blocked CPTH6 effect on LC3BI-II conversion and LC3B puncta formation observed in M14 (Figure 2a) and U-937 cells (Supplementary Figure 1C). In contrast, time course assessment of LC3B turnover demonstrated that bafilomycin A1 or chloroquine failed to further enhance LC3B-II level and LC3B puncta in cells treated with CPTH6 even at shorter time exposure (Figures 2a and b and Supplementary Figure 1E). As expected, the autophagy-related features induced in U-937 cells by canonical stimuli, such as serum starvation, were suppressed by 3-MA (Supplementary Figure 1D), whereas addition of bafilomycin A1 in M14 cells (Figure 2a) and in U-937 (Supplementary Figure 1D) resulted in a conspicuous increase in LC3B-II levels, concomitantly with an increased appearance of puncta EGFP-LC3B. As exposure to 3-MA represents a condition that can paradoxically induce autophagy by inhibiting class I PI3-kinase,19 the effect of 3-MA on Akt/mTOR pathway has been investigated in both U-937 and H1299 cells. Western blot analysis demonstrated that phosphorylation of either Akt at Ser473 or mTOR was not affected by exposure to 3-MA (data not shown).
CPTH6 induces a block of basal autophagy. (a) Quantification of cells positive for autophagosomal structures in M14 cells stably expressing EGFP-LC3B protein after 48 h of treatment with CPTH6 (50 μM), bafilomycin A1 (Baf A1, 2.5 nM), serum starvation or 3-methyladenine (3-MA, 1 mM), alone or in combination. The results represent the average±S.E.M. of three independent experiments. (b) Western blot analysis of LC3B-I to LC3B-II conversion in H1299 cells after treatment with CPTH6 (100 μM) alone or in combination with chloroquine (CQ, 25 μM) or bafilomycin A1 for the indicated times. (c) Representative images of immunofluorescence of p62 protein and (d) quantification of cells positive for p62/EGFP-LC3B colocalization in M14 EGFP-LC3B-expressing cells after CPTH6 treatment (50 μM, 72 h) or serum starvation (48 h). (e) Western blot analysis of p62 protein in U-937 cell lines after serum starvation (48 h). (f) Western blot analysis of NBR1 and p62 proteins in the indicated cell lines after 72 h of treatment with CPTH6. (g) Western blot analysis of HIF-1α, p62, phospho-IkBα (ser34/ser36) and ubiquitin in U-937 cell line after treatment with MG132 (5 μM, 6 h) or CPTH6 (100 μM, 48 h). (b, and e–g) Western blots representative of three independent experiments with similar results are shown. HSP72/73 is shown as loading and transferring control. (b and f) LC3B-II, p62 and NBR1 levels were quantified by densitometric analyses and fold increase relative to untreated cells is presented. (a and d) The results represent the average±S.E.M. of three independent experiments. *P-values were calculated between untreated and treated cells, P<0.05
To monitor the effect of CPTH6 on cargo incorporation, which occurs concurrent with autophagosome maturation, we analyzed colocalization of LC3B protein with the adaptor protein p62.20 As shown in Figures 2c and d, CPTH6 treatment of M14 cells stably expressing EGFP-LC3B induced a large increase in p62 bodies and LC3B puncta when compared with untreated cells. Many of these structures colocalize, and a similar pattern of p62 and LC3B localization was also observed in cells after starvation-induced autophagy (Figures 2c and d). CPTH6 treatment also increased p62 protein level in other tumor cell lines (Figure 2f and Supplementary Figure 2A). An increase in the level of NBR1, another protein selectively degraded during autophagy, was also observed (Figure 2f). As expected, in U-937 cell lines, p62 was reduced by serum starvation (Figure 2e).
As a cross-talk between proteasome-mediated degradation and autophagy has been found,21 and p62 protein degradation may happen via proteasomal pathway,22 we investigated if CPTH6 could act as a proteasome inhibitor. As shown in Figure 2g, increased p62 protein expression after CPTH6 treatment of U-937 cells was associated with modulation of neither ubiquitinated proteins, nor the expression of HIF-1α or phosphorylated IkBα, the two proteins whose degradation proceeds through proteasomal pathway. Moreover, the proteasome inhibitor, MG132, increased the expression of HIF-1α and the phosphorylated IkBα, as expected, whereas it did not affect p62 protein level. Similar results were observed in H1299 cells (Supplementary Figure 2B). Thus, this indicates that in our experimental models, proteasome pathway is not involved in CPTH6-induced p62 protein accumulation.
All together, these results suggest that the increased autophagic markers in CPTH6-treated cells under basal condition are unlikely due to enhanced autophagic flux, but rather by suppression of late maturation stage.
Atg-7, but not Beclin-1, is required for CPTH6-induced autophagic features
Subsequent experiments were performed to evaluate the impact of several proteins acting in different steps of autophagy on the ability of CPTH6 to modulate autophagic markers, such as p62 and LC3B-II. Specifically, the roles of beclin-1-dependent nucleation and Atg-7-mediated elongation steps of the autophagic cascade were investigated, together with the role of LAMP-2, a component of lysosomal membrane proteins, required for the proper fusion of autophagosome and lysosome. To this purpose, H1299 cells were stably transfected with short hairpin RNA (shRNA) targeting beclin-1 or transiently transfected with small interfering (siRNA) directed against Atg-7 or LAMP-2 (Figure 3). As control, cells were transfected with control shRNA vector or scrambled siRNA. Western blot analyses confirmed the effective knockdown of each target expression (Figures 3a–c), and demonstrated that CPTH6 did not affect the expression of beclin-1, Atg-7 and LAMP-2 proteins. Surprisingly, knockdown of beclin-1 failed to prevent the increase in LC3B-II and p62 levels and autophagosome formation triggered by CPTH6 (Figures 3a, d and e), whereas cells depleted of beclin-1 completely prevented starvation-induced autophagy (data not shown). On the contrary, silencing of Atg-7 completely blocked CPTH6 effect on autophagy, being the expression of p62 and LC3B-II and the formation of puncta superimposable to those observed in untreated cells (Figures 3b, d and e). As shown in Figures 3c–e, LAMP-2 silencing resulted in LC3B-II protein accumulation and in a marked increase of EGFP-LC3 puncta compared with untreated cells. On CPTH6 treatment, knockdown of LAMP-2 affected the ability to enhance the expression of p62 and LC3B-II proteins and the formation of puncta.
Atg-7, but not Beclin-1, is required for CPTH6-induced autophagic features. (a–c) Western blot analysis of LC3B-I to LC3B-II conversion, Beclin-1, p62, Atg-7 and LAMP-2 proteins in H1299 cells stably expressing (a) control short hairpin RNA (shCont), short hairpin RNA directed against Beclin-1 (shBECL #1 and #2), or transiently transfected with control RNA interference (siCont), or RNA interference directed against (b) Atg-7 (siAtg-7) or (c) LAMP-2 (siLAMP-2) after CPTH6 treatment (100 μM, 24 h). Western blots representative of three independent experiments with similar results are shown. HSP72/73 is shown as a loading and transferring control. LC3B-II and p62 levels were quantified by densitometric analyses and fold increase relative to untreated cells are presented. (d) Representative images of fluorescence microscopy and (e) quantification of cells positive for autophagosomal structures in H1299 cells stably expressing EGFP-LC3B protein and transiently transfected with shCont, shBECL, siCont, siAtg-7 or siLAMP-2 after treatment with CPTH6 (100 μM, 24 h). The results represent the average±S.E.M. of three independent experiments. P-values were calculated between *untreated and treated cells, or #control (siCont/shCont) and silenced cells, P<0.05
Collectively, these results suggest that CPTH6 triggered an unconventional autophagy that bypassed the beclin-1-dependent nucleation of autophagosomal precursors, and that Atg-7 mediates accelerated autophagic flux resulting from CPTH6 exposure.
Autophagosomes must fuse to lysosomes to degrade their contents. The inability of CPTH6-treated cells to degrade protein aggregates, despite an apparent induction of autophagosomes, prompted us to ask whether CPTH6 interferes with autophagosome–lysosome fusion. To this end, we examined LAMP-2-dependent fusion of autophagosomes with lysosomes to form autolysosomes by tracking the late endosome/lysosome marker LAMP-2, detecting its colocalization with the autophagosomal marker LC3B. As shown in Figure 4a, both serum starvation and CPTH6 treatment induced EGFP-LC3 puncta in M14 cells, but only in CPTH6-treated cells, most of the EGFP-LC3 puncta were not colocalized with LAMP-2, suggesting that fusion of autophagosomes with lysosomes was blocked. On the contrary, under starvation, EGFP-LC3 puncta and LAMP-2 colocalization was observed, as expected, demonstrating the correct fusion between autophagosomes and lysosomes.
CPTH6 blocks autophagy at the late stage. (a) Representative images of immunofluorescence of LAMP-2 protein in M14 EGFP-LC3B-expressing cells after treatment with CPTH6 (50 μM, 72 h) or serum starvation (48 h). (b) Western blot analysis of cathepsin B and cathepsin D proteins in U-937 cells after CPTH6 treatment (48 h). Western blots representative of three independent experiments with similar results are shown. HSP72/73 is shown as a loading and transferring control. (c) Quantification of acid phosphatase activity of U-937 whole-cell extracts or lysosomal fractions obtained after CPTH6 treatment (100 μM, 24 h). The results represent the average±S.D. of two independent experiments and are expressed as optical density (OD) at 540 nm
We also tested whether CPTH6 treatment affected lysosomal function. In particular, the activity of the two lysosomal hydrolases (Figure 4b), cathepsins B and D, that are activated in acidic environment of lysosomes through sequential proteolytic cleavage of their inactive preform, has been evaluated.23 As shown by western blot analysis, using antibodies that recognize both immature and fully mature forms of cathepsin D and cathepsin B, an increased level of the premature form of cathepsin D was detected in U-937 cells after CPTH6 treatment, together with an increase in the heavy chain of the mature enzyme. In contrast, CPTH6 exposure did not cause any significant changes in the expression profile of cathepsin B (Figure 4b).
We next investigated CPTH6 effect on lysosomal permeability. As lysosomal permeability is accompanied by the release of cathepsins into cytosol where they may trigger cellular response, including autophagy, we measured enzymatic activity of acidic phosphatases in both whole-cell extracts and lysosomal fractions. As reported in Figure 4c, CPTH6 treatment did not affect lysosomal permeability activity being the acidic phosphatase similar in untreated and CPTH6-treated cells.
The effect of CPTH6 on autophagy is dependent on ATAT1 expression
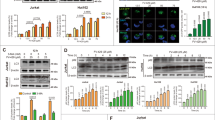
We previously identified CPTH6 as a new inhibitor of histone acetyltransferase activity, and by testing its effect on α-tubulin, a decrease in acetylation level of the protein was observed in U-937 cells14 and H1299 after 48 h of treatment; in contrast, CPTH6 treatment did not modulate the expression of HDAC6 deacetylase (Figure 5a). To dissect the impact of tubulin acetylation status on CPTH6-induced modulation of autophagy, we silenced ATAT1 acetyltransferase and HDAC6 deacetylase expression using the siRNA approach. As shown in Figures 5b, d and e, HDAC6 expression silencing in H1299 cells determined an accumulation of acetylated tubulin, as expected, and concomitantly an induction of LC3B-II accumulation and its recruitment on autophagosome membranes. The same autophagic markers were increased by ATAT1 silencing that, as expected, induced a strong reduction of acetylated tubulin expression (Figures 5c–e). Despite lacking HDAC6 expression, CPTH6 treatment elicited the same effect in terms of induction of LC3B-II accumulation (Figure 5b) and enhancement of percentage of cells with EGFP-LC3B puncta (Figures 5d and e). On the contrary, CPTH6 treatment failed to increase cells with EGFP-LC3B puncta and to accumulate LC3B-II proteins in cells in which the expression of ATAT1 was silenced (Figures 5c–e). Thus, this indicates that the effect of CPTH6 on autophagy is dependent on ATAT1, but not HDAC6, expression.
ATAT1 protein expression is required for CPTH6 effect on autophagy. (a) Western blot analysis of acetylated α-tubulin and HDAC6 proteins in the indicated cell lines after treatment with CPTH6 (100 μM, 48 h). (b and c) Western blot analysis of LC3B-I to LC3B-II conversion, HDAC6 and acetylated α-tubulin proteins in H1299 cells transiently transfected with control RNA interference (siCont), or RNA interference directed against HDAC6 (siHDAC6) or ATAT1 (siATAT1), after treatment with CPTH6 (100 μM, 24 h). LC3B-II levels were quantified by densitometric analyses and fold increase relative to untreated cells is presented. (a–c) Western blots representative of three independent experiments with similar results are shown. HSP72/73 is shown as a loading and transferring control. (d) Representative images of fluorescence microscopy and (e) quantification of cells positive for autophagosomal structures in H1299 cells stably expressing EGFP-LC3B protein and transiently transfected with siHDAC6 or siATAT1 after treatment with CPTH6 (100 μM, 24 h). The results represent the average±S.E.M. of three independent experiments. P-values were calculated between *untreated and treated cells or between #control (siCont) and silenced cells, P<0.05
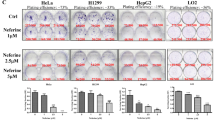
We also assessed whether other compounds having high structural and activity similarities with CPTH6 could produce similar effects on the autophagic pathway. To this aim, U-937 cells were treated with CPTH derivatives CPTH2, CPTH9 and CPTH11 (Supplementary Figure 3A), the three closely related compounds belonging to the same chemical class of CPTH6.13 As reported in Figure 6a, treatment of cells with all CPTH derivatives determined an increase in LC3B-II and p62 proteins expression, even to a different extent. Notably, CPTH2 treatment also induced LC3B puncta in both H1299 (Figures 6b and c) and M14 cells (Supplementary Figure 3B,C) that, similar to CPTH6-treated cells, colocalize with p62 bodies (Figures 6d and e).
CPTH derivatives elicit the same effect of CPTH6 on autophagic flux. (a) Western blot analysis of LC3B-I to LC3B-II conversion and p62 protein in U-937 cell line after treatment with CPTH2 (50 μM, 48 h), CPTH9 or CPTH11 (5 μM, 24 h). Western blots representative of three independent experiments with similar results are shown. HSP72/73 is shown as a loading and transferring control. LC3B-II and p62 levels were quantified by densitometric analyses and fold increase relative to untreated cells is presented. (b) Representative images of fluorescence microscopy and (c) quantification of cells positive for autophagosomal structures in H1299 EGFP-LC3B-expressing cells after 48 h of treatment with CPTH2 (50 μM). (d) Representative images of immunofluorescence of p62 protein and (e) quantification of cells positive for p62/EGFP-LC3B colocalization in M14 EGFP-LC3B-expressing cells after CPTH2 treatment (50 μM, 48 h). (c and e) The results represent the average±S.E.M. of three independent experiments. *P-values were calculated between untreated and treated cells, P<0.05
The effect of CPTH6 on autophagic flux is dependent on the stimulus used to induce autophagy
Previous experiments demonstrated the effects of CPTH6 and its derivatives on basal autophagy. Thus, experiments were also performed to investigate the effect of CPTH6 on autophagy induced by different stimuli, such as serum starvation or RAD001, a second generation of mTOR pharmacological inhibitor. As reported in Figure 7a, H1299 cells exposed to serum starvation or CPTH6 showed an accumulation of LC3B-II compared with untreated cells. On the other hand, cells simultaneously exposed to serum starvation and CPTH6 for 6 h showed no differences in accumulation of LC3-II and p62 when compared with starved or CPTH6-treated cells. To corroborate these results and to test the effect of CPTH6 on autophagosome–lysosome fusion efficiency, we generated H1299 cells stably expressing double-tagged monomeric red fluorescence protein (mRFP)-EGFP-LC3 reporter.24 As shown in Figures 7c and d, we observed a prominent increase in the yellow fluorescence vesicles in CPTH6-treated cells, indicating an incomplete impaired autophagosome maturation in lysosome. On the other hand, serum starvation in the presence or absence of CPTH6 induced the formation of red-only fluorescent vesicles indicative of complete autophagosome maturation. Notably, yellow fluorescence vesicles were observed in starved cells treated with the late-stage inhibitor chloroquine, as expected, and these results were paralleled by the increase of LC3B-II and p62 protein accumulation (Figures 7b–d). As shown in Supplementary Figure 3D,E, CPTH2 treatment of H1299 cells stably expressing mRFP-EGFP-LC3 reporter also led to either a decrease in the percentage of cells with red-only vesicles (mRFP positive) or an increase in the percentage of cells with yellow vesicles (mRFP-EGFP positive).
The effect of CPTH6 on autophagic flux was dependent on the stimulus used to induce autophagy. (a) Western blot analysis of LC3B-I to LC3B-II conversion and p62 protein in H1299 cells after 6 h of treatment with CPTH6 (100 μM) alone or in combination with serum starvation. (b) Western blot analysis of LC3B-I to LC3B-II conversion and p62 protein in H1299 cells after treatment with chloroquine (CQ, 25 μM) alone or in combination with serum starvation (48 h). (c) Representative images of fluorescence microscopy and (d) quantification of cells positive for mRFP (red) and mRFP/GFP (yellow) autophagosomal structures in H1299 cells stably transfected with ptf-LC3B vector after 6 h of treatment with CPTH6 (100 μM) or chloroquine (CQ, 25 μM) alone or in combination with serum starvation. The results represent the average±S.E.M. of three independent experiments. *P-values were calculated between untreated and treated cells, P<0.05. (e) Western blot analysis of LC3B-I to LC3B-II conversion and p62 protein in H1299 cell line after 2 h of treatment with CPTH6 (100 μM) alone or in combination with RAD001 (1 μM). (a, b and e) Western blots representative of three independent experiments with similar results are shown. HSP72/73 is shown as a loading and transferring control. LC3B-II and p62 levels were quantified by densitometric analyses and fold increase relative to untreated cells is presented
Regarding the effect of CPTH6 on RAD001-induced autophagy, both RAD001 and CPTH6 resulted in an enhancement in the LC3B-II levels when compared with untreated cells, whereas, as previously observed (Figure 2f and Supplementary Figure 2A), CPTH6 treatment induced p62 expression and, in combination with RAD001, resulted in a significant increase in the levels of the autophagosomal markers studied (Figure 7e), indicating that the effect of CPTH6 on the autophagic flux was dependent on the stimulus used to induce autophagy.
Discussion
By using different approaches, we demonstrated that CPTH6 inhibits basal autophagy in several human tumor cell lines with different origins, indicating a general effect of CPTH6 instead of a cell type-specific phenomenon.
To analyze which step of autophagosome biogenesis was affected by CPTH6, we used a genetic approach to silence the expression of several proteins acting at different steps of autophagy. Our data indicate that alteration of autophagy in CPTH6-treated cells involved a noncanonical molecular pathway that requires the classic Atg-7 conjugation machinery, but is independent of beclin-1. Beclin-1–p150–Vsp34 complex is considered a core component of the autophagy machinery that drives nucleation; nevertheless, beclin-independent mechanisms of autophagy induction have been demonstrated.25, 26 It is conceivable that autophagosomal biogenesis bypasses the nucleation step, and that Atg proteins were recruited to the pre-existing membranes to drive the subsequent maturation of autophagosomes.
We also demonstrated that CPTH6-induced alterations in the autophagic pathway are LAMP-2 dependent, and LAMP-2 silencing was sufficient to cause the accumulation of LC3B-II and p62 proteins. This evidence is in accord with the notion that (1) LAMP-2 is essential for the proper fusion of lysosomes with autophagosomes, and hence for the removal of autophagosomes and (2) LC3B-II accumulation can be induced by chemical or genetic inhibition of the autophagosome maturation, such as LAMP-227 or rab7 siRNA.28 Moreover, even though CPTH6 treatment induced the expression of cathepsin D, it did not affect the activity of acidic phosphatases and it did not impair lysosomal permeability activity.
The increased number of autophagosomes observed in response to CPTH6 is dependent on the balance between the rate of autophagosome generation and the rate of their conversion into autolysosomes. By inhibiting lysosomal acid hydrolases with bafilomicyn A1 or by preventing endosomal acidification with chloroquine, we determined that autophagosomes did not mature to autophagolysosomes under basal condition, even after short-time exposure to CPTH6, thus indicating that autophagy was blocked at a late stage.
In agreement with these results, p62 and NBR1, two polyubiquitin-binding proteins whose increase in the steady-state levels implies a block in the late stage of autophagy, are enhanced by CPTH6 treatment.20, 24 Even though p62 protein degradation may happen via proteasomal pathway,22 and a cross-talk between proteasome-mediated degradation and autophagy has been demonstrated,21 we have evidence that CPTH6 did not act as a proteasome inhibitor.
Furthermore, inhibition of autophagic flux by CPTH6 is most likely a general function of CPTH derivatives and does not represent a cell type-specific phenomenon, as other three compounds with similar chemical structure caused the same inhibitory effect in several human tumor cell lines of different origins.
α-Tubulin has been identified as a major ATAT1/MEC-17 acetyltransferase29 and HDAC6 deacetylase30 substrate. The evidence that, in our experimental models, downregulation of both HDAC6 and ATAT1 increases markers of autophagy corroborates previous results demonstrating that regulation of acetylation/deacetylation balance of α-tubulin has important implications for autophagy.31 As CPTH6 inhibits α-tubulin acetylation, our hypothesis is that CPTH6 could affect autophagosome maturation through its effect on α-tubulin acetylation. Tubulin hyperacetylation has been demonstrated to be necessary to allow kinesin-1-dependent JNK activation and stimulation of autophagosome formation,32 and hence a possible effect of CPTH6 on this pathway can be hypothesized. Moreover, the evidence that CPTH6 treatment failed to increase cells with EGFP-LC3B puncta and to accumulate p62 and LC3B-II proteins in cells in which the expression of ATAT1 was silenced led us to speculate that ATAT1 could be another direct target of CPTH6; indeed, our previously published results identified CPTH6 as a novel pCAF and Gcn5 histone acetyltransferase inhibitor14 and ATAT1 just belongs to the Gcn5 histone acetyltransferase family.33 Nevertheless, we cannot exclude that CPTH6 may induce changing in tubulin acetylation status interfering with the activity of other acetyltransferases (like the Elongator complex ELP3 subunit)34 and/or deacetylase such as SIRT2.35 These data are in agreement with the evidence that defective autolysosome maturation after CPTH6 treatment was observed.
As, unlike what happens with tubulin, Atg proteins, including Atg-5, Atg-7 and Atg-8, need to be deacetylated to stimulate autophagy, in a way that involves p300 and SIRT1,36, 37 we cannot exclude that CPTH6 could also act through an impairment of these proteins.
Our results also demonstrated that the effect of CPTH6 on autophagy was dependent on the stimulus used to induce autophagy. In fact, a different response of tumor cells, in terms of LC3B-II induction, was observed when the effect of CPTH6 was evaluated on nutrient-independent basal autophagy, or autophagy induced by serum starvation, or by pharmacological inducers such as RAD001. This difference can be related to different autophagic modalities previously demonstrated by different groups. For instance, HDAC6 has been demonstrated to control autophagosome maturation in basal autophagy condition, but to be completely dispensable for starvation-induced autophagy.38 Similarly, kinesin-1 has been demonstrated to carry autophagosomes in basal condition, but it is not involved in motoring autophagosomes after nutrient deprivation.32 Indeed, the decrease of autophagosome mobility associated with tubulin hyperacetylation induced by serum starvation32 can be responsible for the different responses of cells to CPTH6 treatment in terms of enhancement of autophagic markers observed in the presence or absence of serum.
Taken together, our results expand our current knowledge about the bioactivity of CPTH6 and strongly suggest that CPTH6 acts on multiple targets in cancer cells and affects two different mechanisms: apoptosis and autophagy. Our previous paper demonstrated the ability of CPTH6 to activate the apoptotic program in several cancer cell lines.14 In the present study we have demonstrated the ability of CPTH6 to prevent autophagy completion. The observation that either bcl-2 and bcl-xL overexpression or co-treatment with a pan-caspase inhibitor did not prevent CPTH6-induced LC3B conversion (data not shown) strongly suggests that the effect of CPTH6 on autophagic flux is independent of the ability of the compound to induce apoptosis. Dual targeting of apoptosis and autophagy by the same agent is not a new concept: inhibitors of Bcl-2,39 Hsp90,40 AMPK41 and proteasome42 have been demonstrated to induce both apoptosis and autophagy in cancer cells.
In conclusion, CPTH6 could be a valuable agent for the treatment of cancer and should be further developed as a potent anticancer agent, not only by inducing apoptosis but also by affecting autophagy.
Materials and Methods
Cell cultures and plasmids
Human AML (HL-60, U-937), melanoma (M14), lung (H1299, NCI-H460) and ovary (OVCA 433, SKOV-3) carcinoma cell lines were cultured in 10% inactivated fetal bovine serum (HyClone, Thermo Scientific, South Logan, UT, USA) in RPMI medium (Invitrogen, Carlsbad, CA, USA). For starvation, cells were incubated in RPMI medium without any supplement. Human ovary and lung carcinoma cell lines were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). AML cell lines were a generous gift from Dr. Mauro Valtieri (Istituto Superiore di Sanità, Rome, Italy). Cells were stably transfected using Lipofectamine2000 (Invitrogen) according to the manufacturer’s instructions. For EGFP-LC3B, mRFP-EGFP-LC3B (ptfLC3, plasmid 21074; Addgene, Cambridge, MA, USA), shBeclin-1 and sh-Control expressing cells, stable clones or mixed populations were cultured in the presence of geneticin (800 μg/ml, Sigma-Aldrich, St. Louis, MO, USA).
Pooled siRNA oligonucleotides (siGENOME SMART pool) against Atg-7, LAMP-2 and ATAT1 were purchased from Dharmacon RNA Technologies (Lafayette, CO, USA). For siRNA transfection, cells were seeded and transfected with 100 nM pooled oligonucleotide mixture by using Lipofectamine 2000 following the manufacturer’s protocol. After transfection, media were removed and cells were allowed to recover in complete growth media for 48 h before using them for experiments. Gene silencing efficacy by various shRNAs or siRNAs was assessed by western blot assay.
Reagent preparation and treatments
CPTH6 and its analogs, cyclopentylidene-[4-(4′-chlorophenyl)thiazol-2-yl)hydrazone (CPTH2), 1-(4-(4-methoxyphenyl)thiazol-2-yl)-2-(2-methylcyclohexylidene)hydrazone (CPTH9) and 1-(4-(4-methoxyphenyl)thiazol-2-yl)-2-(3-methylcyclohexylidene)hydrazone (CPTH11), were synthesized and dissolved as previously reported.13, 14 For all experiments, cells were treated with 1% DMSO as control. After 24 h from seeding, exponentially growing AML and solid tumor cells were treated with CPTH6 at concentrations ranging from 20 to 100 μM for 6–72 h. Bafilomicyn A1 (Baf A1, Santa Cruz Biotechnology, Santa Cruz, CA, USA) and 3-MA (Enzo Life Science, Plymouth Meeting, PA, USA), mTOR inhibitor RAD001 (GlaxoSmithKline, Brentford, Middlesex, UK) and proteasome inhibitor Z-leu-leu-leu-CHO (MG132, Santa Cruz Biotechnology) were dissolved in DMSO. Chloroquine diphosphate (CQ, Sigma-Aldrich) was dissolved in water.
Transmission electron microscopy
After treatment, cells were collected and centrifuged; the pellet was fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.3) at room temperature for 20 min. After postfixation with 1% OsO4 in 0.2 M cacodylate buffer (pH 7.3) at room temperature for 30 min, cells were dehydrated with ascending concentrations of ethanol, and embedded in epoxy resin (TAAB Laboratories Equipment Limited, Aldermarton, UK). Ultrathin sections, obtained with an LKB Ultratome Nova ultramicrotome (LKB, Bromma, Sweden), were stained with uranyl acetate and lead citrate and were examined with a Philips EM 208S electron microscope (FEICo, Eindhoven, The Netherlands).
Western blot analysis
After treatments, both detached and adherent cells were collected, lysed and total protein extracts were fractionated by SDS-PAGE, transferred to a nitrocellulose filter and subjected to immunoblot assay, as previously described.14 Immunodetection was performed using antibodies directed to: p62, LAMP-2, HDAC6, NBR1, Ubiquitin (Santa Cruz Biotechnology), HSP72/73 (Calbiochem, San Diego, CA, USA), LC3B, acetyl-α-tubulin (K40; Sigma-Aldrich), cathepsin B, cathepsin D (Calbiochem), Atg-7 (Millipore, Billerica, MA, USA), phospho-Ik-Bα (ser32/ser36), total and phospho (ser473) AKT, total and phospho (ser2448) mTOR (Cell Signaling, Danvers, MA, USA) and HIF-1α (BD Pharmigen, San Diego, CA, USA). Anti-mouse or anti-rabbit immunoglobulin G (IgG)-horseradish peroxidase conjugated antibodies (Cell Signaling; Amersham Biosciences, Freiburg, Germany) were used as secondary antibodies in a 1 : 10 000 dilution. Antibody binding was visualized by enhanced chemiluminescence method (Amersham Biosciences) according to the manufacturer’s specification and recorded on autoradiographic film (Amersham Biosciences).
Densitometric analysis
Developed films were acquired using GS-700 Imaging Densitometer (Bio-Rad Laboratories, Hercules, CA, USA) and processed with Corel Photo Paint 7.0 (Corel inc., Mountain View, CA, USA) to adjust image brightness and contrast. Densitometric evaluation was performed using Molecular Analyst Software (Bio-Rad) and normalized with relative controls depending on the analysis. In particular, LC3B-II and p62 protein levels versus housekeeping protein expression were analyzed and fold increase relative to control was presented.
Immunofluorescence microscopy
Cells grown on glass coverslips were fixed in 2% formaldehyde for 10 min at room temperature. Detection of autophagosomal structures was performed by fluorescence microscopy observing LC3B puncta in EGFP-LC3B-expressing cells. Autophagic flux was analyzed by fluorescence microscopy monitoring the distribution and alteration of mRFP-GFP-LC3B fluorescent signals. Colocalization experiments were performed using EGFP-LC3B M14-expressing cells stained with primary antibodies directed to p62 or LAMP-2 (Santa Cruz Biotechnology). Tetramethylrhodamine-5-(and 6)-isothiocyanate-conjugated Goat anti-mouse antibody (Jackson Lab, Bar Harbor, ME, USA) was used as a secondary antibody. Typically, at least 200 cells were counted, and cells with more than 10 puncta were considered autophagy positive.
For all fluorescence microscopy experiments, images were scanned under a × 63 oil immersion objective and, to avoid bleed-through effects, each fluorescent signal was scanned independently using a Leica DMIRE2 microscope equipped with a Leica DFC 350FX camera, elaborated by a Leica FW4000 deconvolution software (Leica, Solms, Germany) and processed using Adobe PhotoShop software (Adobe Inc., Burlington, NJ, USA).
Lysosomal subfractionation and acid phosphatase assay
After treatments, cell homogenates from ∼3 × 108 cells were enriched for lysosomal fraction by differential centrifugation followed by density centrifugation (Lysosome Extraction kit, Sigma-Aldrich). Acid phosphatase activity in homogenates was evaluated by Acid Phosphatase Assay kit (Sigma-Aldrich) according to the manufacturer’s protocol.
Statistics
Experiments were replicated three times, unless otherwise indicated, and the data were expressed as means±S.D. or mean±S.E.M. Differences between groups were analyzed with a two-sided paired or unpaired t-test and they were considered to be statistically significant for P<0.05.
Abbreviations
- CPTH6:
-
3-methylcyclopentylidene-[4-(4′-chlorophenyl)thiazol-2-yl]hydrazone
- 3-MA:
-
3-methyladenine
- EGFP:
-
enhanced green fluorescence protein
- RFP:
-
red fluorescence protein
- LC3B:
-
microtubule-associated protein 1 light chain 3
- LAMP-2:
-
lysosome-associated membrane protein 2
References
Wirawan E, Vanden Berghe T, Lippens S, Agostinis P, Vandenabeele P . Autophagy: for better or for worse. Cell Res 2012; 22: 43–61.
Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006; 441: 885–889.
Eisenberg-Lerner A, Bialik S, Simon HU, Kimchi A . Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ 2009; 16: 966–975.
Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev 2010; 90: 1383–1435.
Weidberg H, Shvets E, Elazar Z . Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem 2011; 80: 125–156.
Das G, Shravage BV, Baehrecke EH . Regulation and function of autophagy during cell survival and cell death. Cold Spring Harbor Perspect Biol 2012; 4: a008813.
Kimura S, Noda T, Yoshimori T . Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct Funct 2008; 33: 109–122.
Ganley IG, Wong PM, Gammoh N, Jiang X . Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell 2011; 42: 731–743.
Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y . Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct 1998; 23: 33–42.
Carew JS, Kelly KR, Nawrocki ST . Autophagy as a target for cancer therapy: new developments. Cancer Manag Res 2012; 4: 357–365.
Firat E, Weyerbrock A, Gaedicke S, Grosu AL, Niedermann G . Chloroquine or chloroquine-PI3K/Akt pathway inhibitor combinations strongly promote γ-irradiation-induced cell death in primary stem-like glioma cells. PLoS One 2012; 7: e47357.
Baek KH, Park J, Shin I . Autophagy-regulating small molecules and their therapeutic applications. Chem Soc Rev 2012; 41: 3245–3263.
Chimenti F, Bizzarri B, Maccioni E, Secci D, Bolasco A, Chimenti P et al. A novel histone acetyltransferase inhibitor modulating Gcn5 network: cyclopentylidene-[4-(4′-chlorophenyl)thiazol-2-yl)hydrazone. J Med Chem 2009; 52: 530–536.
Trisciuoglio D, Ragazzoni Y, Pelosi A, Desideri M, Carradori S, Gabellini C et al. CPTH6, a thiazole derivative, induces histone hypoacetylation and apoptosis in human leukemia cells. Clin Cancer Res 2012; 18: 475–486.
Gordy C, He YW . The crosstalk between autophagy and apoptosis: where does this lead? Protein Cell 2012; 3: 17–27.
Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 2008; 4: 151–175.
Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8: 445–544.
Tanida I . Autophagosome formation and molecular mechanism of autophagy. Antioxid Redox Signal 2011; 14: 2201–2214.
Wu YT, Tan HL, Shui G, Bauvy C, Huang Q, Wenk MR et al. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem 2010; 285: 10850–10861.
Kirkin V, McEwan DG, Novak I, Dikic I . A role for ubiquitin in selective autophagy. Mol Cell 2009; 34: 259–269.
Lamark T, Johansen T . Autophagy: links with the proteasome. Curr Opin Cell Biol 2010; 22: 192–198.
Lee J, Kim HR, Quinley C, Kim J, Gonzalez-Navajas J, Xavier R et al. Autophagy suppresses interleukin-1β (IL-1β) signaling by activation of p62 degradation via lysosomal and proteasomal pathways. J Biol Chem 2012; 287: 4033–4040.
Ishidoh K, Kominami E . Processing and activation of lysosomal proteinases. Biol Chem 2002; 383: 1827–1831.
Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282: 24131–24145.
Codogno P, Mehrpour M, Proikas-Cezanne T . Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol 2011; 13: 7–12.
Seo G, Kim SK, Byun YJ, Oh E, Jeong SW, Chae GT et al. Hydrogen peroxide induces Beclin 1-independent autophagic cell death by suppressing the mTOR pathway via promoting the ubiquitination and degradation of Rheb in GSH-depleted RAW 264.7 cells. Free Radic Res 2011; 45: 389–399.
Fortunato F, Bürgers H, Bergmann F, Rieger P, Büchler MW, Kroemer G et al. Impaired autolysosome formation correlates with Lamp-2 depletion: role of apoptosis, autophagy, and necrosis in pancreatitis. Gastroenterology 2009; 137: 350–360.
Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E . Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 2005; 1: 84–91.
Castro-Castro A, Janke C, Montagnac G, Paul-Gilloteaux P, Chavrier P . ATAT1/MEC-17 acetyltransferase and HDAC6 deacetylase control a balance of acetylation of alpha-tubulin and cortactin and regulate MT1-MMP trafficking and breast tumor cell invasion. Eur J Cell Biol 2012; 91: 950–960.
Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A et al. HDAC6 is a microtubule-associated deacetylase. Nature 2002; 417: 455–458.
Xie R, Nguyen S, McKeehan WL, Liu L . Acetylated microtubules are required for fusion of autophagosomes with lysosomes. BMC Cell Biol 2010; 11: 89.
Geeraert C, Ratier A, Pfisterer SG, Perdiz D, Cantaloube I, Rouault A et al. Starvation-induced hyperacetylation of tubulin is required for the stimulation of autophagy by nutrient deprivation. J Biol Chem 2010; 285: 24184–24194.
Akella JS, Wloga D, Kim J, Starostina NG, Lyons-Abbott S, Morrissette NS et al. MEC-17 is an alpha-tubulin acetyltransferase. Nature 2010; 467: 218–222.
Creppe C, Malinouskaya L, Volvert ML, Gillard M, Close P, Malaise O et al. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell 2009; 136: 551–564.
North BJ, Marshall BL, Borra MT, Denu JM, Verdin E . The human Sir2 ortholog, SIRT2, is an NAD -dependent tubulin deacetylase. Mol Cell 2003; 11: 437–444.
Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA 2008; 105: 3374–3379.
Lee IH, Finkel T . Regulation of autophagy by the p300 acetyltransferase. J Biol Chem 2009; 284: 6322–6328.
Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J 2010; 29: 969–980.
Niu X, Li S, Wei F, Huang J, Wu G, Xu L et al. Apogossypolone induces autophagy and apoptosis in breast cancer MCF-7 cells in vitro and in vivo. Breast Cancer 2012 doi:10.1007/s12282-012-0372-z.
Liu KS, Liu H, Qi JH, Liu QY, Liu Z, Xia M et al. SNX-2112, an Hsp90 inhibitor, induces apoptosis and autophagy via degradation of Hsp90 client proteins in human melanoma A-375 cells. Cancer Lett 2012; 318: 180–188.
Yang WL, Perillo W, Liou D, Marambaud P, Wang P . AMPK inhibitor compound C suppresses cell proliferation by induction of apoptosis and autophagy in human colorectal cancer cells. J Surg Oncol 2012; 106: 680–688.
Choi CH, Lee BH, Ahn SG, Oh SH . Proteasome inhibition-induced p38 MAPK/ERK signaling regulates autophagy and apoptosis through the dual phosphorylation of glycogen synthase kinase 3beta. Biochem Biophys Res Commun 2012; 418: 759–764.
Acknowledgements
We are grateful to Dr. Ameeta Kelekar and Dr. Jonathan Howard for providing shBeclin-1, and EGFP-LC3B plasmids, respectively, Dr. Adele Petricca for secretarial assistance and Dr. Simone Bonacelli for English revision. Dr. Ylenia Ragazzoni and Dr. Teresa De Luca are recipients of a fellowship from the Italian Foundation for Cancer Research. This work was supported by grants from the Italian Association for Cancer Research (MFAG 11502 (to DT); IG 10239 (to DDB); and 5 × 1000, 9979 (to DDB)).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Stephanou
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Ragazzoni, Y., Desideri, M., Gabellini, C. et al. The thiazole derivative CPTH6 impairs autophagy. Cell Death Dis 4, e524 (2013). https://doi.org/10.1038/cddis.2013.53
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.53
Keywords
This article is cited by
-
The α-tubulin acetyltransferase ATAT1: structure, cellular functions, and its emerging role in human diseases
Cellular and Molecular Life Sciences (2024)
-
Pharmacological targeting of CBP/p300 drives a redox/autophagy axis leading to senescence-induced growth arrest in non-small cell lung cancer cells
Cancer Gene Therapy (2023)
-
Acetaldehyde Induces Cytotoxicity via Triggering Mitochondrial Dysfunction and Overactive Mitophagy
Molecular Neurobiology (2022)
-
Inhibition of lysine acetyltransferases impairs tumor angiogenesis acting on both endothelial and tumor cells
Journal of Experimental & Clinical Cancer Research (2020)
-
Bcl-2-dependent synthetic lethal interaction of the IDF-11774 with the V0 subunit C of vacuolar ATPase (ATP6V0C) in colorectal cancer
British Journal of Cancer (2018)