Abstract
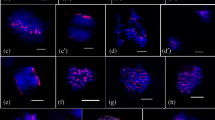
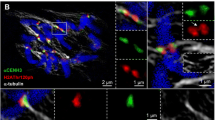
Histone acetylation affects chromatin conformation and transcriptional activity. However, the structural role of histone acetylation at specific chromosomal regions, such as the centromere, is poorly understood. In this study, histone H4 acetylation and its localization in barley interphase nuclei are revealed by three-dimensional microscopy. The centromeres form a ring-like allocation near the nuclear membrane in barley. Immunofluorescence studies on non-fixed, interphase nuclei treatment revealed ring-like distribution of the highly acetylated histone H4, located near the nuclear membrane at one pole of the nucleus. This fluorescent structure was similar to the centromere cluster and referred to as hyperacetylated region (HAR). The distribution pattern of the acetylated histone H4 was similar to each of the K5, K8, K12 and K16 lysine residues, although H4 acetylated at K5, K8 and K12 residues was found in almost all nuclei, whereas H4 acetylated at K16 was weakly observed in only half of the nuclei. Each HAR consists of two strongly acetylated cores and a halo-like, less acetylated surrounding area. Fluorescence signals from centromere-specific repetitive sequences of barley, detected through three-dimensional fluorescence in situ hybridization (3D-FISH), co-localized with the HAR corresponding to the K5 residue acetylation, but the signals did not completely overlap each other. These findings indicate that histone acetylation specifically occurring at the centromeres likely have certain structural roles for the centromere.
Similar content being viewed by others
References
Agard, D.A., Hiraoka, Y., Shaw, P. and Sedat, J.W. 1989. Fluorescence microscopy in three dimensions. Meth. Cell Biol. 30: 353–377.
Annunziato, A.T., Frabo, L.Y., Seale, R.L. and Woodcock, C.L.F. 1988 Treatment with sodium butyrate inhibits the complete condensation of interphase chromatin. Chromosoma 96: 132–138.
Aragón-Alcaide, L., Miller, T., Schwarzacher, T., Reader, S. and Moore, G. 1996. A cereal centromeric sequence. Chromosoma 105: 261–268.
Arumuganathan, K. and Earle, E.D. 1991. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9: 208–218.
Belyaev, N.D., Houben, A., Baranczewski, P. and Schubert, I. 1997. Histone H4 acetylation in plant heterochromatin is altered during the cell cycle. Chromosoma 106: 193–197.
Belyaev, N.D., Houben, A., Baranczewski, P. and Schubert, I. 1998. The acetylation patterns of histone H3 and H4 along Vicia faba chromosomes are different. Chromosome Res. 6: 59–63.
Brownell, J.E. and Allis, C.D. 1996. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr. Opin. Genet. Dev. 6: 176–184.
Comings, D.E. 1980 Arrangement of chromatin in the nucleus. Hum. Genet. 53: 131–143.
Cremer, T., Cremer, C., Baumann, H., Luedtke, E.K., Sperling, K., Teuber, V. and Zorn, C. 1982. Rabl's model of the interphase chromosome arrangement tested in Chinese hamster cells by premature chromosome condensation and laser-UV-microbeam experiments. Hum. Genet. 60: 46–56.
Cremer, T., Dietzel, S., Eils, R., Lichter, P. and Cremer, C. 1995. Chromosome territories, nuclear matrix filaments and interchromatin channels: a topological view on nuclear architecture and function. In: P.E. Brandham and M.D. Bennett (Eds.) Kew Chromosome Conference IV, Kew Royal Botanic Gardens, pp. 63–81.
Dawe, R.K., Reed, L.M., Yu, H., Muszynski, M.G. and Hiatt, E.N. 1999. A maize homolog of mammalian CENPC is a constitutive component of the inner kinetochore. Plant Cell 11: 1227–1238.
Dong, F. and Jiang, J. 1998. Non-Rabl patterns of centromere and telomere distribution in the interphase nuclei of plant cells. Chromosome Res. 6: 551–558.
Fukui, K. and Kakeda, K. 1990 Quantitative karyotyping of barley chromosomes by image analysis methods. Genome 33: 450–458.
Fukui, K. 1996. Plant Chromosomes: Laboratory Methods. CRC Press, Boca Raton, FL, p. 1.
Funabiki, H., Hagan, I., Uzawa, S. and Yanagida, M. 1993. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 121: 961–976.
Garcia-Ramirez, M., Rocchini, C. and Ausio, J. 1995. Modulation of chromatin folding by histone acetylation. J. Biol. Chem. 270: 17923–17928.
Hendzel, M.J., Sun, J., Chen, H.Y., Rattner, J.B. and Davie, J.R. 1994. Histone acetyltransferase is associated with the nuclear matrix. J. Biol. Chem. 269: 22894–22901.
Houben, A., Guttenbach, M. and Schubert, I. 1995. Nucleolus-like bodies in field bean cells. Biol. Zentralbl. 114: 339–345.
Houben, A., Belyaev, N.D., Leach, C.R. and Timmis, J.N. 1997. Differences of histone H4 acetylation and replication timing between A and B chromosomes Brachycome dichromosomatica. Chromosome Res. 5: 233–237.
Iwano, M., Fukui, K., Takaichi, S. and Isogai, A. 1997. Globular and fibrous structure in barley chromosomes revealed by highresolution scanning microscopy. Chromosome Res. 5: 341–349.
Jasencakova, Z., Meister, A., Walter, J., Turner, B.M. and Schubert, I. 2000. Histone H4 acetylation of euchromatin and heterochromatin is cell cycle dependent and correlated with replication rather than with transcription. Plant Cell 12: 2087–2100.
Jasencakova, Z., Meister, A. and Schubert, I. 2001. Chromatin organization and its relation to replication and histone acetylation during the cell cycle in barley. Chromosoma 110: 83–92.
Jeppesen, P. and Turner, B.M. 1993. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell 74: 281–289.
Jiang, J., Nasuda, S., Dong, F., Scherrer, C.W., Woo, S., Wing, R.A., Gill, B.S. and Ward, D.C. 1996. A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc. Natl. Acad. Sci. USA 93: 14210–14213.
Jones, P.L., Veenstra, G.J.C., Wade, P.A., Vermaak, D., Kass, S.U., Landsberger, N., Stroubouli, J. and Wolffe, A.P. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nature Genet. 19: 187–191.
Kumekawa, N., Ohmido, N., Fukui, K., Otsubo, E. and Otsubo, H. 2001. A new gypsy-type retrotransposon RIRE7: preferential insertion into the tandem repeat sequence TrsD in the pericentromeric heterochromatin regions of rice chromosomes. Mol. Gen. Genet. 265: 480–488.
Künzel, G., Korzun, L. and Meister, A. 2000. Cytologically integrated physical restriction fragment length polymorphism maps for the barley genome based on translocation breakpoints. Genetics 154: 397–412.
Kuo, M.H., Brownell, J.E., Sobel, R.E., Ranalli, T.A., Cook, R.G., Edmonson, D.G., Roth, S.Y. and Allis, C.D. 1996. Transcriptionlinked acetylation by Gcn5p of histone H3 and H4 at specific lysines. Nature 383: 269–272.
Luger, K., Mäder, A.W., Richmond, R.K., Sargent, D.F. and Richmond, T.J. 1997. Crystal structure of the nucleosome core particle at 2.8Å resolution. Nature 389: 251–260.
Moroi, Y., Hartman, A.L., Nakane, P.K., Tan, E.M. 1981. Distribution of kinetochore (centromere) antigen in mammalian cell nuclei. J. Cell Biol. 90: 245–259.
Nan, X., Ng, H., Johnson, C.A., Laherty, C.D., Turner, B.M., Eisenman, R.N. and Bird, A. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393: 386–388.
Ng, H.H. and Bird, A. 2000. Histone deacetylases: silencers for hire. Trends Biol. Sci. 25: 121–126.
Noguchi, J. and Fukui, K. 1995. Chromatin arrangements in intact interphase nuclei examined by laser confocal microscopy. J. Plant Res. 108: 209–216.
Ohmido, N. and Fukui, K. 1997. Visual verification of close disposition between a rice A genome-specific DNA sequence (TrsA) and the telomere sequence. Plant Mol. Biol. 35: 963–968.
Ohmido, N., Kijima, K., Ashikawa, I., de Jong, J.H. and Fukui, K. 2001 Visualization of the terminal structure of rice chromosomes 6 and 12 using multicolor FISH to chromosomes and extended DNA fibers. Plant Mol. Biol. 47: 413–421.
Presting, G., Malysheva, L., Fuchs, J. and Schubert, I. 1998. A TY3/GYPSY retrotransposon-like sequence localizes to the centromeric regions of cereal chromosomes. Plant J. 16: 721–728.
Richards, E.J. and Ausubel, F.M. 1988. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell 53: 127–136.
Selig, S., Okumura, K., Ward, D.C. and Cedar, H. 1992. Delineation of DNA replication time zones by fluorescence in situ hybridization. EMBO J. 11: 1217–1225.
Sommerville, J., Baird, J. and Turner, B.M. 1993. Histone H4 acetylation and transcription in amphibian chromatin. J. Cell Biol. 120: 277–290.
Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12: 599–606.
Sugimoto, K., Fukuda, R. and Himeno, M. Centromere/kinetochore localization of human centromere protein A (CENP-A) exogenously expressed as a fusion to green fluorescent protein. Cell Struct. Funct. 25: 253–261.
Turner, B.M. and Fellows, G. 1989. Specific antibodies reveal ordered and cell-cycle-related use of histone-H4 acetylation sites in mammalian cells. Eur. J. Biochem. 179: 131–139.
Turner, B.M., Birley, A.J. and Lavender, J. 1992. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell 69: 375–384.
Vourc'h, C., Taruscio, D., Boyle, A.L. and Ward, D.C. 1993. Cell cycle-dependent distribution of telomeres, centromeres, and chromosome-specific subsatellite domains in the interphase nucleus of mouse lymphocytes. Exp. Cell Res. 205: 142–151.
Wako, T., Fukuda, M., Furushima-Shimogawara, R., Belyaev, N.D., Turner, B.M. and Fukui, K. 1998. Comparative analysis of topographic distribution of acetylated histone H4 by using confocal microscopy and a deconvolution system. Anal. Chim. Acta 365: 9–17.
Wako, T., Fukuda, M., Furushima-Shimogawara, R., Belyaev, N.D. and Fukui, K. Cell cycle dependent and lysine residue-specific dynamic changes of histone H4 acetylation in barley. Plant Mol. Biol. 46: 645–653.
White, D.A., Belyaev, N.D. and Turner, B.M. 1999. Preparation of site-specific antibodies to acetylated histones. Methods 19: 417–421.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wako, T., Houben, A., Furushima-Shimogawara, R. et al. Centromere-specific acetylation of histone H4 in barley detected through three-dimensional microscopy. Plant Mol Biol 51, 533–541 (2003). https://doi.org/10.1023/A:1022375017938
Issue Date:
DOI: https://doi.org/10.1023/A:1022375017938