Abstract
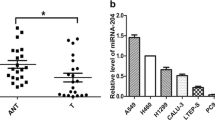
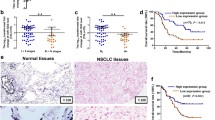
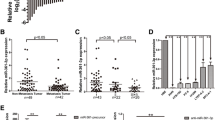
MicroRNAs (miRNAs) as a class of small noncoding RNA molecules regulate the expression of targeted gene. The dysregulation of microRNAs is reported to be involved in carcinogenesis and tumor progression. Here, we identified miR-140-3p as a downregulated microRNA in most cancer tissues including lung cancer tissues, compared with their normal counterparts. MiR-140-3p was observed to perform its tumor suppressor function via its inhibition on cell growth, migration and invasion but its induction of cell apoptosis. Furthermore, the growth of non-small-cell lung cancer (NSCLC) cells in nude mouse models were suppressed by overexpression of miR-140-3p. ATP8A1 was demonstrated as a novel direct target of miR-140-3p using a luciferase assay. The increased level of intracellular ATP8A1 protein attenuated the inhibitor role of miR-140-3p in the growth and mobility of NSCLC cell. A regulation mechanism of miR-140-3p for the development and progression of NSCLC through downregulating the ATP8A1 expression was first discovered in the present study.






Similar content being viewed by others
References
Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA: A Cancer J Clini. 2011;61:212–36.
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: A Cancer J Clin. 2011;61:69–90.
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA: A Cancer J Clin. 2012;62:10–29.
Asmis TR, Ding K, Seymour L, Shepherd FA, Leighl NB, Winton TL, et al. Age and comorbidity as independent prognostic factors in the treatment of non small-cell lung cancer: A review of national cancer institute of canada clinical trials group trials. J Clin Oncol: Off J Ame Soc Clin Oncol. 2008;26:54–9.
Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, et al. Cancer statistics, 2005. CA: A Cancer J Clin. 2005;55:10–30.
Bartel DP. Micrornas: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Ambros V. The functions of animal micrornas. Nature. 2004;431:350–5.
Zanetti KA, Haznadar M, Welsh JA, et al. 3′-UTR and functional secretor haplotypes in mannose-binding lectin 2 are associated with increased colon cancer risk in African Americans. Cancer Res. 2012;72(6):1467–77.
Kirigin FF et al. Dynamic microRNA gene transcription and processing during T cell development. J Immunol. 2012;188:3257–67.
Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–48.
Macrae IJ et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–8.
Lau PW, Guiley KZ, De N, Potter CS, Carragher B, MacRae IJ. The molecular architecture of human Dicer. Nat Struct Mol Biol. 2012;19:436–40.
Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–82.
Calin GA, Croce CM. Microrna signatures in human cancers. Nat Rev Cancer. 2006;6:857–66.
Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14.
Tu Y et al. MicroRNA-218 inhibits glioma invasion, migration, proliferation, and cancer stem-like cell self-renewal by targeting the polycomb group gene Bmi1. Cancer Res. 2013;73(19):6046–55.
Hui W et al. MicroRNA-195 inhibits the proliferation of human glioma cells by directly targeting cyclin D1 and cyclin E1. PLoS One. 2013;8(1), e54932.
Guan S et al. Overexpressed miRNA-137 inhibits human glioma cells growth by targeting Rac1. Cancer Biother Radiopharm. 2013;28(4):327–34.
Yang TQ et al. MicroRNA-16 inhibits glioma cell growth and invasion through suppression of BCL2 and the nuclear factor-kappaB1/MMP9 signaling pathway. Cancer Sci. 2014;105(3):265–71.
Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microrna host genes and transcription units. Genome Res. 2004;14:1902–10.
Tardif G, Pelletier JP, Fahmi H, Hum D, Zhang Y, Kapoor M, et al. Nfat3 and tgf-beta/smad3 regulate the expression of mir-140 in osteoarthritis. Arthritis Res Ther. 2013;15:R197.
Sand M, Skrygan M, Sand D, Georgas D, Hahn SA, Gambichler T, et al. Expression of micrornas in basal cell carcinoma. Br J Dermatol. 2012;167:847–55.
Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. Microrna signatures in human ovarian cancer. Cancer Res. 2007;67:8699–707.
Tan X, Qin W, Zhang L, Hang J, Li B, Zhang C, et al. A 5-microrna signature for lung squamous cell carcinoma diagnosis and hsa-mir-31 for prognosis. Clin Cancer Res: Off J Am Assoc Cancer Res. 2011;17:6802–11.
Song B, Wang Y, Xi Y, Kudo K, Bruheim S, Botchkina GI, et al. Mechanism of chemoresistance mediated by mir-140 in human osteosarcoma and colon cancer cells. Oncogene. 2009;28:4065–74.
Kai Y, Peng W, Ling W, Jiebing H, Zhuan B. Reciprocal effects between microrna-140-5p and adam10 suppress migration and invasion of human tongue cancer cells. Biochem Biophys Res Commun. 2014;448:308–14.
Yang H, Fang F, Chang R, Yang L. Microrna-140-5p suppresses tumor growth and metastasis by targeting transforming growth factor beta receptor 1 and fibroblast growth factor 9 in hepatocellular carcinoma. Hepatology. 2013;58:205–17.
Yuan Y, Shen Y, Xue L, Fan H. Mir-140 suppresses tumor growth and metastasis of non-small cell lung cancer by targeting insulin-like growth factor 1 receptor. PLoS One. 2013;8, e73604.
Usui A, Hoshino I, Akutsu Y, Sakata H, Nishimori T, Murakami K, et al. The molecular role of fra-1 and its prognostic significance in human esophageal squamous cell carcinoma. Cancer. 2012;118:3387–96.
Hu H, Li S, Liu J, Ni B. Microrna-193b modulates proliferation, migration, and invasion of non-small cell lung cancer cells. Acta Biochim Biophys Sin. 2012;44:424–30.
Chen LT, Xu SD, Xu H, Zhang JF, Ning JF, Wang SF. Microrna-378 is associated with non-small cell lung cancer brain metastasis by promoting cell migration, invasion and tumor angiogenesis. Med Oncol. 2012;29:1673–80.
Pai LM, Barcelo G, Schupbach T. D-cbl, a negative regulator of the EGFR pathway, is required for dorsoventral patterning in Drosophila oogenesis. Cell. 2000;103:51–61.
Ashkenazi A. Targeting death and decoy receptors of the tumour-necrosis factor superfamily. Nat Rev Cancer. 2002;2:420–30.
Soupene E, Kuypers FA. Identification of an erythroid atp-dependent aminophospholipid transporter. Br J Haematol. 2006;133:436–8.
Soupene E, Kemaladewi DU, Kuypers FA. Atp8a1 activity and phosphatidylserine transbilayer movement. J Recept Ligand Channel Res. 2008;1:1–10.
Kato U, Inadome H, Yamamoto M, Emoto K, Kobayashi T, Umeda M. Role for phospholipid flippase complex of ATP8A1 and CDC50A proteins in cell migration. J Biol Chem. 2013;288:4922–34.
Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microrna molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98.
Bandi N, Zbinden S, Gugger M, Arnold M, Kocher V, Hasan L, et al. Mir-15a and mir-16 are implicated in cell cycle regulation in a rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009;69:5553–9.
Takahashi Y, Forrest AR, Maeno E, Hashimoto T, Daub CO, Yasuda J. Mir-107 and mir-185 can induce cell cycle arrest in human non small cell lung cancer cell lines. PLoS One. 2009;4, e6677.
Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A microrna signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–801.
Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- rna genes mir15 and mir16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–9.
Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microrna expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61.
Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, et al. Specific activation of microrna-127 with downregulation of the proto-oncogene bcl6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–43.
O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. C-myc-regulated micrornas modulate e2f1 expression. Nature. 2005;435:839–43.
Zhang Y, Eades G, Yao Y, Li Q, Zhou Q. Estrogen receptor alpha signaling regulates breast tumor-initiating cells by down-regulating mir-140 which targets the transcription factor sox2. J Biol Chem. 2012;287:41514–22.
Daleke DL, Lyles JV. Identification and purification of aminophospholipid flippases. Biochim Biophys Acta. 2000;1486:108–27.
Levano K, Punia V, Raghunath M, Debata PR, Curcio GM, Mogha A, et al. Atp8a1 deficiency is associated with phosphatidylserine externalization in hippocampus and delayed hippocampus-dependent learning. J Neurochem. 2012;120:302–13.
Fadok VA, de Cathelineau A, Daleke DL, Henson PM, Bratton DL. Loss of phospholipid asymmetry and surface exposure of phosphatidylserine is required for phagocytosis of apoptotic cells by macrophages and fibroblasts. J Biol Chem. 2001;276:1071–7.
Martin SJ, Reutelingsperger CP, McGahon AJ, Rader JA, van Schie RC, LaFace DM, et al. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of bcl-2 and abl. J Exp Med. 1995;182:1545–56.
Fadok VA, Bratton DL, Rose DM, Pearson A, Ezekewitz RA, Henson PM. A receptor for phosphatidylserine-specific clearance of apoptotic cells. Nature. 2000;405:85–90.
Acknowledgments
This work was support by The development of science and technology plan projects of Chinese medicine in Shandong, No. 2013-209.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dong, W., Yao, C., Teng, X. et al. MiR-140-3p suppressed cell growth and invasion by downregulating the expression of ATP8A1 in non-small cell lung cancer. Tumor Biol. 37, 2973–2985 (2016). https://doi.org/10.1007/s13277-015-3452-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-3452-9