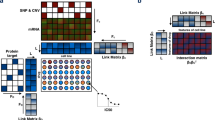
Abstract
Radiation and hormone level targeted drug therapy are one of the most widely adopted treatment options for different types of cancer. But, due to genetic variations, cancer patients shows heterogeneity towards targeted drug therapies. In such a scenario precision medication necessitates the design of targeted drug therapy for each individual based on their genetic structure. Predictive modeling and drug response data of cancer cells are imperative in designing personalized cancer treatment. Recent advancement in cancer research has produced various pharmacogenomic databases, which further encourages ongoing research in precision medication. In this paper, we have proposed the drug sensitivity prediction framework using ensemble and multi-task learning. The proposed framework successfully maps non-linear relationships among anti-cancer drugs and have modeled their dependency. Further, the proposed framework is validated using publicly available real data sets-GDSC, CCLE, NCI-Dream. The proposed ensemble model shows quite promise in predicting anti-cancer drug response and has achieved lesser mean square error 3.28 (CGP), 0.49 (CCLE) and 0.54 (NCI-DREAM) in comparison to other existing counterparts.


Similar content being viewed by others
References
Argyriou A, Evgeniou T, Pontil M (2008) Convex multi-task feature learning. Mach Learn 73(3):243–272
Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D et al (2012) The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483(7391):603–607
Brubaker D, Difeo A, Chen Y, Pearl T, Zhai K, Bebek G, Chance M, Barnholtz-Sloan J (2014) Drug intervention response predictions with paradigm (DIRPP) identifies drug resistant cancer cell lines and pathway mechanisms of resistance. In: Pacific symposium on biocomputing. pacific symposium on biocomputing. NIH Public Access, p 125
Cameron A (2015) Targeted braf inhibitors: immunological effects and combination with immunotherapy. http://hdl.handle.net/10063/4937
Conti JA, Kemeny NE, Saltz LB, Huang Y, Tong WP, Chou T-C, Sun M, Pulliam S, Gonzalez C (1996) Irinotecan is an active agent in untreated patients with metastatic colorectal cancer. J Clin Oncol 14(3):709–715
Costello JC, Heiser LM, Georgii E, Gönen M, Menden MP, Wang NJ, Bansal M, Hintsanen P, Khan SA, Mpindi J-P et al (2014) A community effort to assess and improve drug sensitivity prediction algorithms. Nat Biotechnol 32(12):1202–1212
Cuzick J (1985) A wilcoxon-type test for trend. Stat Med 4(4):543–547
Daemen A, Griffith OL, Heiser LM, Wang NJ, Enache OM, Sanborn Z, Pepin F, Durinck S, Korkola JE, Griffith M et al (2013) Modeling precision treatment of breast cancer. Genome Biol 14(10):110–124
Evgeniou T, Pontil M (2004) Regularized multi-task learning. In: Proceedings of the tenth ACM SIGKDD international conference on Knowledge discovery and data mining. ACM, pp 109–117
Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J et al (2012) Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483(7391):570–575
Garraway LA (2013) Genomics-driven oncology: framework for an emerging paradigm. J Clin Oncol 31(15):1806–1814
Gönen M, Margolin AA (2014) Drug susceptibility prediction against a panel of drugs using kernelized bayesian multitask learning. Bioinformatics 30(17):i556–i563
Gupta S, Chaudhary K, Kumar R, Gautam A, Nanda JS, Dhanda SK, Brahmachari SK, Raghava GPS (2016) Prioritization of anticancer drugs against a cancer using genomic features of cancer cells: a step towards personalized medicine. Sci Rep 6:23857
He X, Cai D, Niyogi P (2006) Laplacian score for feature selection. In: Advances in neural information processing systems, pp 507–514
Heider D, Senge R, Cheng W, Hüllermeier E (2013) Multilabel classification for exploiting cross-resistance information in HIV-1 drug resistance prediction. Bioinformatics 29(16):1946–1952
Jie H, Shaozhi F, Peng Q, Han Y, Xie J, Zan N, Chen Y, Fan J (2017) Paclitaxel-loaded polymeric nanoparticles combined with chronomodulated chemotherapy on lung cancer: in vitro and in vivo evaluation. Int J Pharm 516(1):313–322
Jang IS, Neto EC, Guinney J, Friend SH, Margolin AA (2014) Systematic assessment of analytical methods for drug sensitivity prediction from cancer cell line data. In: Pacific symposium on biocomputing. NIH Public Access, p 63
Kantarjian HM, Giles FJ, Bhalla KN, Pinilla-Ibarz JA, Larson RA, Gattermann N, Ottmann OG, Hochhaus A, Radich JP, Saglio G et al (2010) Nilotinib is effective in patients with chronic myeloid leukemia in chronic phase following imatinib resistance or intolerance: 24-month follow-up results. Blood 117(4):1141–1145
MacConaill LE, Garraway LA (2010) Clinical implications of the cancer genome. J Clin Oncol 28(35):5219–5228
Menden MP, Iorio F, Garnett M, McDermott U, Benes CH, Ballester PJ, Saez-Rodriguez J (2013) Machine learning prediction of cancer cell sensitivity to drugs based on genomic and chemical properties. PLoS One 8(4):e61318
National Center for Health Statistics US et al (2015) Health, United States, 2014: with special feature on adults aged, pp 55–64
Neto EC, Jang IS, Friend SH, Margolin AA (2014) The stream algorithm: computationally efficient ridge-regression via Bayesian model averaging, and applications to pharmacogenomic prediction of cancer cell line sensitivity. In: Pacific symposium on biocomputing. NIH Public Access, pp 27–38
Paul R, Hawkins SH, Balagurunathan Y, Schabath MB, Gillies RJ, Hall LO, Goldgof DB (2016) Deep feature transfer learning in combination with traditional features predicts survival among patients with lung adenocarcinoma. Tomography 2(4):388
Radovic M, Ghalwash M, Filipovic N, Obradovic Z (2017) Minimum redundancy maximum relevance feature selection approach for temporal gene expression data. BMC Bioinform 18(1):9
Rhee S-Y, Taylor J, Wadhera G, Ben-Hur A, Brutlag DL, Shafer RW (2006) Genotypic predictors of human immunodeficiency virus type 1 drug resistance. Proc Natl Acad Sci 103(46):17355–17360
Sánchez-Maroño N, Alonso-Betanzos A, Tombilla-Sanromán M (2007) Filter methods for feature selection—a comparative study. In: International conference on intelligent data engineering and automated learning. Springer, pp 178–187
Sharma A, Rani R (2017a) Classification of cancerous profiles using machine learning. In: 2017 international conference on machine learning and data science (MLDS). IEEE, pp 31–36
Sharma A, Rani R (2017b) An optimized framework for cancer classification using deep learning and genetic algorithm. J Med Imaging Health Inform 7(8):1851–1856
Sharma A, Rani R (2018a) C-HDESHO: cancer classification framework using single objective meta-heuristic and machine learning approaches. In: 2018 fifth international conference on parallel, distributed and grid computing (PDGC). IEEE, pp 406–411
Sharma A, Rani R (2018b) BE-DTI’: ensemble framework for drug target interaction prediction using dimensionality reduction and active learning. Comput Methods Programs Biomed 165:151–162
Sharma A, Rani R (2018c) An integrated framework for identification of effective and synergistic anti-cancer drug combinations. J Bioinform Comput Biol 16(05):1850017
Sharma A, Rani R (2018d) KSRMF: kernelized similarity based regularized matrix factorization framework for predicting anti-cancer drug responses. J Intell Fuzzy Syst 35(2):1779–1790
Sharma A, Rani R (2019) C-HMOSHSSA: gene selection for cancer classification using multi-objective meta-heuristic and machine learning methods. Comput Methods Programs Biomed 178:219–235
Sheng J, Li F, Wong STC (2015) Optimal drug prediction from personal genomics profiles. IEEE J Biomed Health Inform 19(4):1264–1270
Shoemaker RH (2006) The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer 6(10):813–823
Wan Q, Pal R (2014) An ensemble based top performing approach for NCI-dream drug sensitivity prediction challenge. PLoS One 9(6):e101183
Wang H, Cao Q, Dudek AZ (2012) Phase II study of panobinostat and bortezomib in patients with pancreatic cancer progressing on gemcitabine-based therapy. Anticancer Res 32(3):1027–1031
Wolpert DH (1992) Stacked generalization. Neural Netw 5(2):241–259
Zhang N, Wang H, Fang Y, Wang J, Zheng X, Liu XS (2015) Predicting anticancer drug responses using a dual-layer integrated cell line-drug network model. PLoS Comput Biol 11(9):e1004498
Zsebik B, Citri A, Isola J, Yarden Y, Szöllősi J, Vereb G (2006) Hsp90 inhibitor 17-aag reduces erbb2 levels and inhibits proliferation of the trastuzumab resistant breast tumor cell line jimt-1. Immunol Lett 104(1):146–155
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sharma, A., Rani, R. Drug sensitivity prediction framework using ensemble and multi-task learning. Int. J. Mach. Learn. & Cyber. 11, 1231–1240 (2020). https://doi.org/10.1007/s13042-019-01034-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13042-019-01034-0