Abstract
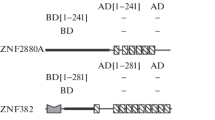
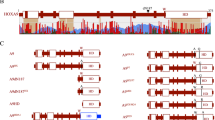
We previously characterized a C2H2-type zinc finger protein HZF1 (ZNF16) and demonstrated its important role in erythroid and megakaryocytic differentiation. This protein was located in nucleus. In this study, we first approved that HZF1 solely could activate lacZ reporter gene in yeast host Y190. This self-activation phenomenon together with structure and distribution of HZF1 suggested it as a potential transcription factor. By the auto-activation experiments and the luciferase reporter system and deletion mutation analysis, we further located the trans-activation domain at amino acid residences 49–197 within the non-zinc finger region of HZF1. An acidic residue-rich subregion (amino acids 49–105) was important for the trans-activation effect, but it could not function independently. By deletion mutation analysis, we also identified three nuclear location signals, which were located in the regions of amino acids 255–280, 328–360, and 460–490, respectively, and all of them within the zinc finger region.




Similar content being viewed by others
References
Miller, J., McLachlan, A. D., & Klug, A. (1985). Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. The EMBO Journal, 4, 1609–1614.
Lee, M. S., Gippert, G. P., Soman, K. V., Case, D. A., & Wright, P. E. (1989). Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science, 245, 635–637.
Moore, M., & Ullman, C. (2003). Recent developments in the engineering of zinc finger proteins. Briefings in Functional Genomics and Proteomics, 1, 342–355.
Shivdasani, R. A., & Orkin, S. H. (1996). The transcriptional control of hematopoiesis. Blood, 87, 4025–4039.
Shivdasani, R. A. (1997). Stem cell transcription factors. Hematology/Oncology Clinics of North America, 11, 1199–1206.
Fujiwara, Y., Browne, C. P., Cunniff, K., Goff, S. C., & Orkin, S. H. (1996). Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proceedings of the National Academy of Sciences of the United States of America, 93, 12355–12358.
Chen, D., & Zhang, G. (2001). Enforced expression of the GATA-3 transcription factor affects cell fate decisions in hematopoiesis. Experimental Hematology, 29, 971–980.
Perry, C., & Soreq, H. (2002). Transcriptional regulation of erythropoiesis: Fine tuning of combinatorial multi-domain elements. European Journal of Biochemistry, 269, 3607–3618.
Hock, H., & Orkin, S. H. (2006). Zinc-finger transcription factor Gfi-1: versatile regulator of lymphocytes, neutrophils and hematopoietic stem cells. Current Opinion in Hematology, 13, 1–6.
Molnár, A., Wu, P., Largespada, D. A., Vortkamp, A., Scherer, S., Copeland, N. G., et al. (1996). The Ikaros gene encodes a family of lymphocyte-restricted zinc finger DNA binding proteins, highly conserved in human and mouse. Journal of Immunology, 156, 585–592.
McCarty, A. S., Kleiger, G., Eisenberg, D., & Smale, S. T. (2003). Selective dimerization of a C2H2 zinc finger subfamily. Molecules and Cells, 11, 459–470.
Georgopoulos, K., Winandy, S., & Avitahl, N. (1997). The role of the Ikaros gene in lymphocyte development and homeostasis. Annual Review of Immunology, 15, 155–176.
Wang, J. H., Nichogiannopoulou, A., Wu, L., Sun, L., Sharpe, A. H., Bigby, M., et al. (1996). Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity, 5, 537–549.
Peng, H., Du, Z. W., & Zhang, J. W. (2006). Identification and characterization of a novel zinc finger protein (HZF1) gene and its function in erythroid and megakaryocytic differentiation of K562 cells. Leukemia, 20, 1109–1116.
Ruden, D. M. (1992). Activating regions of yeast transcription factors must have both acidic and hydrophobic amino acids. Chromosoma, 101, 342–348.
Ruden, D. M., Ma, J., Li, Y., Wood, K., & Ptashne, M. (1991). Generating yeast transcriptional activators containing no yeast protein sequences. Nature, 350, 250–252.
Triezenberg, S. J. (1995). Structure and function of transcriptional activation domains. Current Opinion in Genetics & Development, 5, 190–196.
Mitchell, P. J., & Tjian, R. (1989). Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science, 245, 371–378.
Johnson, P. F., Sterneck, E., & Williams, S. C. (1993). Activation domains of transcriptional regulatory proteins. The Journal of Nutritional Biochemistry, 4, 386–398.
Attardi, L. D., & Tjian, R. (1993). Drosophila tissue-specific transcription factor NTF-1 contains a novel isoleucine-rich activation motif. Genes and Development, 7, 1341–1353.
Chen, X., & Bieker, J. J. (1996). Erythroid Krüppel-like factor (EKLF) contains a multifunctional transcriptional activation domain important for inter- and intramolecular interactions. The EMBO Journal, 15, 5888–5896.
Choy, B., & Green, M. R. (1994). Eukaryotic activators function during multiple steps of preinitiation complex assembly. Nature, 366, 531–536.
Kim, T. K., & Roeder, R. (1994). Proline-rich activator CTF1 targets the TFIIB assembly step during transcriptional activation. Proceedings of the National Academy of Sciences of the United States of America, 91, 4170–4174.
Xiao, H., Pearson, A., Coulombe, B., Truat, R., Zhang, S., Regier, J. L., et al. (1994). Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Molecular and Cellular Biology, 14, 7013–7024.
Lüscher, B., & Eisenman, R. N. (1990). New light on myc and myb. Part II. myb. Genes and Development, 4, 2235–2241.
Hupp, T. R., Meek, D. W., Midgley, C. A., & Lane, D. P. (1992). Regulation of the specific DNA binding function of p53. Cell, 71, 875–886.
Nerlov, C., & Ziff, E. B. (1994). Three levels of functional interaction determine the activity of CCAAT/enhancer binding protein-alpha on the serum albumin promoter. Genes and Development, 8, 350–362.
Mattaj, I. W., & Englmeier, L. (1998). Nucleocytoplasmic transport: the soluble phase. Annual Review of Biochemistry, 67, 265–306.
Schiestl, R. H., & Gietz, R. D. (1989). High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Current Genetics, 6, 339–346.
Chien, C. T., Bartel, P. L., Sternglanz, R., & Fields, S. (1991). The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proceedings of the National Academy of Sciences of the United States of America, 88, 9578–9582.
Acknowledgments
This work was supported by National Nature Science Foundation of China (30870532 and 30721063) and the Special Fund of National Laboratory of China (2060204).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Supplement: Detection of nuclear location signals (NLSs) within HZF1 are shown below.
Supplement Fig. 1
Cellular localization of GFP fused with HZF1 fragment in HeLa cells transfected with partial recombination constructs. (A) DAPI staining of the HeLa cells transfected with the constructs expressing GFP fused a different HZF1 fragment. The nuclei were displayed in bright blue under fluorescent microscope at the wavelength 365 nm excitation. (B) The expression of GFP fused a HZF1 fragment in the transfected HeLa cells was observed under fluorescent microscope at the wavelength 488 nm excitation. It is noticeable that only partial cells were transformed and expressed GFP. (C) The combined result of (A) and (B) to highlight the location of GFP fused HZF1 fragment. (D) The corresponding HZF1 fragments that were fused with GFP in the recombination constructs used for the transfection experiments. (JPG 257 kb)
Supplement Fig. 2
Cellular localization of GFP fused with HZF1 fragment in HeLa cells transfected with partial recombination constructs. (A) DAPI staining of the HeLa cells transfected with the constructs expressing GFP fused a different HZF1 fragment. The nuclei were displayed in bright blue under fluorescent microscope at the wavelength 365 nm excitation. (B) The expression of GFP fused a HZF1 fragment in the transfected HeLa cells was observed under fluorescent microscope at the wavelength 488 nm excitation. It is noticeable that only partial cells were transformed and expressed GFP. (C) The combined result of (A) and (B) to highlight the location of GFP fused HZF1 fragment. (D) The corresponding HZF1 fragments that were fused with GFP in the recombination constructs used for the transfection experiments. (JPG 265 kb)
Supplement Fig. 3
Cellular localization of GFP fused with HZF1 fragment in HeLa cells transfected with partial recombination constructs. (A) DAPI staining of the HeLa cells transfected with the constructs expressing GFP fused a different HZF1 fragment. The nuclei were displayed in bright blue under fluorescent microscope at the wavelength 365 nm excitation. (B) The expression of GFP fused a HZF1 fragment in the transfected HeLa cells was observed under fluorescent microscope at the wavelength 488 nm excitation. It is noticeable that only partial cells were transformed and expressed GFP. (C) The combined result of (A) and (B) to highlight the location of GFP fused HZF1 fragment. (D) The corresponding HZF1 fragments that were fused with GFP in the recombination constructs used for the transfection experiments. (JPG 314 kb)
Supplement Fig. 4
Cellular localization of GFP fused with HZF1 fragment in HeLa cells transfected with partial recombination constructs. (A) DAPI staining of the HeLa cells transfected with the constructs expressing GFP fused a different HZF1 fragment. The nuclei were displayed in bright blue under fluorescent microscope at the wavelength 365 nm excitation. (B) The expression of GFP fused a HZF1 fragment in the transfected HeLa cells was observed under fluorescent microscope at the wavelength 488 nm excitation. It is noticeable that only partial cells were transformed and expressed GFP. (C) The combined result of (A) and (B) to highlight the location of GFP fused HZF1 fragment. (D) The corresponding HZF1 fragments that were fused with GFP in the recombination constructs used for the transfection experiments. (JPG 267 kb)
Rights and permissions
About this article
Cite this article
Deng, MJ., Li, XB., Peng, H. et al. Identification of the Trans-Activation Domain and the Nuclear Location Signals of Human Zinc Finger Protein HZF1 (ZNF16). Mol Biotechnol 44, 83–89 (2010). https://doi.org/10.1007/s12033-009-9210-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-009-9210-8