Abstract
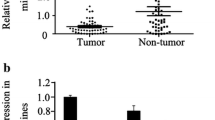
There has been few report discussing the expression and function of miR-212 in gastric cancer (GC). The aim of this pilot study was to investigate the expression of miR-212 in both gastric cancer tissues and gastric cancer cells and further explores the possible reasons for this change and the impact on the development of gastric cancer. qRT–PCR was used to detect the expression of miR-212 in primary GC tissues, adjacent normal tissues, gastric cancer cell lines BGC-823, SGC-7901, MKN-45, and normal gastric mucosa cell line GES. The expression of miR-212 was evaluated before and after treatment with methylation inhibitor-5-Aza-2′-deoxycitidine (5-Aza-dC), finally anti-miRNA and dual luciferase reporter assay were used to prove that MYC is a target gene of miR-212. The results showed that a significant reduction of miR-212 expression in GC tissues was observed compared to that in normal tissues (P = 0.002). At the same time, miR-212 expression level in normal gastric mucosa cell line GES was higher than that of in gastric cancer cell lines BGC-823, SGC-7901, and MKN-45 (P = 0.015, 0.008, 0.044, respectively). Computer sequence analysis showed the hypermethylation of CpG islands(CPI) in the promoter regions of miR-212 led to the lower expression of miR-212 in gastric cell strains (BGC-823 and SGC-7901). MiR-212 expression was significantly recovered after treatment with methylation inhibitor 5-Aza-dC (P = 0.016, 0.000, 0.015, respectively). Then, the results of AMOs transfection and dual luciferase reporter assay showed that Myc is a target of miR-212, which will be helpful to verify the function of miR-212 in carcinogenesis. The conclusion could be deduced from the study that decreased expression of miR-212 may be due to hypermethylation of CPI in gastric cancer cells, and miR-212 might act on the progression of gastric cancer through the potential target gene Myc.





Similar content being viewed by others
References
Visone R, Petrocca F, Croce CM. Micro-RNAs in gastrointestinal and liver disease. Gastroenterology. 2008;135(6):1866–9.
Garzon R, Fabbri M, Cimmino A, et al. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12(12):580–7.
Chivukula RR, Mendell JT. Circular reasoning: microRNAs and cell-cycle control. Trends Biochem Sci. 2008;33(10):474–81.
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–66.
Esquela-Kerscher A, Slack FJ. Oncomirs- microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–69.
Yang L. Incidence and mortality of gastric cancer in China. World J Gastroentrol. 2006;12(1):17–20.
Chen CF, Ridzon DA, Broomer AJ. Real- time quantification of microRNAs by stem-loop RT- PCR. Nucleic Acids Res. 2005;33(20):e179.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using Real-Time quantitative PCR and the 2−∆∆Ct method. Methods. 2001;25(4):402–8.
Kaneda A, Kaminishi M, Sugimura T, et al. Decreased expression of the seven ARP2/3 complex genes in human gastric cancers. Cancer Lett. 2004;212(2):203–10.
Lehmann U, Hasemeier B, Römermann D, et al. Epigenetic inactivation of microRNA genes in mammary carcinoma. Verh Dtsch Ges Pathol. 2007;91:214–20.
Weber B, Stresemann C, Brueckner B, et al. Methylation of human microRNA genes in normal and neoplastic cells. Cell Cycle. 2007;6(9):1001–5.
Kozaki K, Imoto I, Moqi S, et al. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68(7):2094–105.
Yang N, Coukos G, Zhang L. MicroRNA epigenetic alterations in human cancer: one step forward in diagnosis and treatment. Int J Cancer. 2008;122(5):963–8.
Ando T, Yoshida T, Enomoto S, et al. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int J Cancer. 2009;124(10):2367–74.
Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol. 2006;16(4):318–30.
van Waardenburg RC, Meijer C, Pinto-Sietsma SJ, et al. Effects of c-myc oncogene modulation on differentiation of human small cell lung carcinoma cell lines. Anticancer Res. 1998;18(1A):91–5.
Robson S, Pelengaris S, Khan M. c-Myc and downstream targets in the pathogenesis and treatment of cancer. Recent Pat Anticancer Drug Discov. 2006;1(3):305–26.
Acknowledgments
We thank Miss Yong Hua for excellent advice and critical reading of this paper.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xu, L., Wang, F., Xu, XF. et al. Down-regulation of miR-212 expression by DNA hypermethylation in human gastric cancer cells. Med Oncol 28 (Suppl 1), 189–196 (2011). https://doi.org/10.1007/s12032-010-9691-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12032-010-9691-0