Abstract
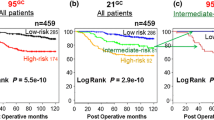
We recently developed a 95-gene classifier (95GC) for the prognostic prediction for ER-positive and node-negative breast cancer patients treated with only adjuvant hormonal therapy. The aim of this study was to validate the efficacy of 95GC and compare it with that of 21GC (Oncotype DX) as well as to evaluate the combination of 95GC and 21GC. DNA microarray data (gene expression) of ER-positive and node-negative breast cancer patients (n = 459) treated with adjuvant hormone therapy alone as well as those of ER-positive breast cancer patients treated with neoadjuvant chemotherapy (n = 359) were classified with 95GC and 21GC (Recurrence Online at http://www.recurrenceonline.com/). 95GC classified the 459 patients into low-risk (n = 285; 10 year relapse-free survival: 88.8 %) and high-risk groups (n = 174; 70.6 %) (P = 5.5e−10), and 21GC into low-risk group (n = 286; 89.3 %), intermediate-risk (n = 81; 75.7 %), and high-risk (n = 92; 64.7 %) groups (P = 2.9e−10). The combination of 95GC and 21GC classified them into low-risk (n = 324; 88.9 %) and high-risk (n = 135; 65.0 %) groups (P = 5.9e−14), and also showed that pathological complete response rates were significantly (P = 2.5e−6) higher for the high-risk (17.9 %) than the low-risk group (3.6 %). In addition, we demonstrated that 95GC was calculated on a single-sample basis if the reference robust multi-array average workflow was used for normalization. The prognostic prediction capability of 95GC appears to be comparable to that of 21GC. Moreover, their combination seems to result in the identification of more low-risk patients who do not need chemotherapy than either classification alone. The patients in the high-risk group were found to be more chemo-sensitive so that they can benefit more from adjuvant chemotherapy.



Similar content being viewed by others
Abbreviations
- ER:
-
Estrogen receptor
- PR:
-
Progesterone receptor
- HER2:
-
Human epidermal growth factor receptor 2
References
Sparano JA, Paik S (2008) Development of the 21-gene assay and its application in clinical practice and clinical trials. J Clin Oncol 26:721–728
Prat A, Ellis MJ, Perou CM (2011) Practical implications of gene-expression-based assays for breast oncologists. Nat Rev Clin Oncol 9:48–57
Prat A, Perou CM (2011) Deconstructing the molecular portraits of breast cancer. Mol Oncol 5:5–23
Paik S (2011) Is gene array testing to be considered routine now? Breast 20(3):S87–S91
Perou CM, Borresen-Dale AL (2011) Systems biology and genomics of breast cancer. Cold Spring Harb Perspect Biol 3(2):a003293. doi:10.1101/cshperspect.a003293
Kim C, Paik S (2010) Gene-expression-based prognostic assays for breast cancer. Nat Rev Clin Oncol 7:340–347
Ross JS, Hatzis C, Symmans WF et al (2008) Commercialized multigene predictors of clinical outcome for breast cancer. Oncolog 13:477–493
Colombo PE, Milanezi F, Weigelt B, Reis-Filho JS (2011) Microarrays in the 2010s: the contribution of microarray-based gene expression profiling to breast cancer classification, prognostication and prediction. Breast Cancer Res 13:212
Gokmen-Polar Y, Badve S (2012) Molecular profiling assays in breast cancer: are we ready for prime time? Oncolog (Williston Park) 26:350–357, 361
Naoi Y, Kishi K, Tanei T et al (2011) Development of 95-gene classifier as a powerful predictor of recurrences in node-negative and ER-positive breast cancer patients. Breast Cancer Res Treat 128:633–641
Paik S (2007) Development and clinical utility of a 21-gene recurrence score prognostic assay in patients with early breast cancer treated with tamoxifen. Oncologist 12:631–635
Harris L, Fritsche H, Mennel R et al (2007) American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol 25:5287–5312
NCCN Clinical Practice Guidelines in Oncology, Breast Cancer (Version 1.2011). http://www.nccn.org/professionals/physician_gls/f_guidelines.asp
Sparano JA (2006) TAILORx: trial assigning individualized options for treatment (Rx). Clin Breast Cancer 7:347–350
Paik S, Shak S, Tang G et al (2004) A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351:2817–2826
Gyorffy B, Benke Z, Lanczky A et al (2012) RecurrenceOnline: an online analysis tool to determine breast cancer recurrence and hormone receptor status using microarray data. Breast Cancer Res Treat 132:1025–1034
Tsunashima R, Naoi Y, Kishi K et al (2012) Estrogen receptor positive breast cancer identified by 95-gene classifier as at high risk for relapse shows better response to neoadjuvant chemotherapy. Cancer Lett 324:42–47
Naoi Y, Kishi K, Tanei T et al (2011) Prediction of pathologic complete response to sequential paclitaxel and 5-fluorouracil/epirubicin/cyclophosphamide therapy using a 70-gene classifier for breast cancers. Cancer 117:3682–3690
Naoi Y, Tanei T, Kishi K et al (2012) 70-Gene classifier for differentiation between paclitaxel- and docetaxel-sensitive breast cancers. Cancer Lett 314:206–212
Parker JS, Mullins M, Cheang MC et al (2009) Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 27:1160–1167
Morimoto K, Kim SJ, Tanei T et al (2009) Stem cell marker aldehyde dehydrogenase 1-positive breast cancers are characterized by negative estrogen receptor, positive human epidermal growth factor receptor type 2, and high Ki67 expression. Cancer Sci 100:1062–1068
Elston CW, Ellis IO (1991) Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19:403–410
Goldstein DR (2006) Partition resampling and extrapolation averaging: approximation methods for quantifying gene expression in large numbers of short oligonucleotide arrays. Bioinformatics 22:2364–2372
Katz S, Irizarry RA, Lin X et al (2006) A summarization approach for Affymetrix GeneChip data using a reference training set from a large, biologically diverse database. BMC Bioinformatics 7:464
Irizarry RA, Hobbs B, Collin F et al (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264
Chang JC, Makris A, Gutierrez MC et al (2008) Gene expression patterns in formalin-fixed, paraffin-embedded core biopsies predict docetaxel chemosensitivity in breast cancer patients. Breast Cancer Res Treat 108:233–240
Symmans WF, Hatzis C, Sotiriou C et al (2010) Genomic index of sensitivity to endocrine therapy for breast cancer. J Clin Oncol 28:4111–4119
Acknowledgments
This study was supported, in part, by the Knowledge Cluster Initiative of the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Conflict of interest
SN received honoraria and research funding from Sysmex Corp. KK and YB are employees of Sysmex Corp.
Ethical standards
The study complied with the current laws of Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Naoi, Y., Kishi, K., Tsunashima, R. et al. Comparison of efficacy of 95-gene and 21-gene classifier (Oncotype DX) for prediction of recurrence in ER-positive and node-negative breast cancer patients. Breast Cancer Res Treat 140, 299–306 (2013). https://doi.org/10.1007/s10549-013-2640-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-013-2640-9