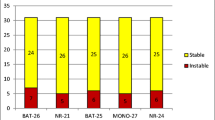
Abstract
The colorectal cancer harbor germline, somatic or epimutations in mismatch repair genes, MUTYH or POLE gene, which lead to the hypermutated and ultramutator phenotypes with increased immune response. The mutations in POLE gene were reported to occur more frequently in early-onset colorectal cancer (EOCRC), and the patients are strong candidates for checkpoint inhibitor therapy. Here, we report mutation analysis within the endonuclease domain of the POLE gene in the cohort of patients with EOCRC in order to identify recurrent or new mutations and evaluate their association with the presence of tumor-infiltrating lymphocytes (TILs) and peritumoral lymphoid reaction. We have shown a significant association between MSI tumors and TILs (p = 0.004). Using sensitive single-tube nested PCR with subsequent Sanger sequencing, we have found in one female patient diagnosed at age 48 with rectal adenocarcinoma with mucinous elements staged pT3pN2pM1 a silent variant within the exon 9 NM_006231.3 c.849 C > T, NP_00622.2 p.Leu283 = recorded in dSNP as rs1232888774 with MAF = 0.00002. In silico prediction, result showed possible involvement into splicing; therefore, this rare variant can be involved into EOCRC pathogenesis. In the time of precise medicine, it is important to develop screening strategies also for less common conditions such as EOCRC allowing to predict tailored therapy for younger patients suffering from CRC that harbor mutations in the POLE gene.



Similar content being viewed by others
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Network Cancer Genome Atlas. Comprehensive molecular characterization of human and rectal cancer. Nature. 2012;18:330–7.
Mahasneh A, Al-Shaheri F, Jamal E. Molecular biomarkers for an early diagnosis, effective treatment and prognosis of colorectal cancer: current updates. Exp Mol Pathol. 2017;102:475–83.
Pilati C, Shinde J, Alexandrov LB, et al. Mutational signature analysis identifies MUTYH deficiency in colorectal cancers and adrenocortical carcinomas. J Pathol. 2017;242:10–5.
Cohen R, Sroussi M, Pilati C, Houry S, Laurent-Puig P, André T. Unresectable metastatic colorectal cancer patient cured with cetuxomab-based chemotherapy: a case report with new molecular insights. Gastrointest Oncol. 2018;9:E23–7.
Palles C, Cazier JB, Howarth KM, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45:136–44.
Bellido F, Pineda M, Aiza G, et al. POLE and POLD1 mutations in 529 kindred with familial colorectal cancer and/or polyposis: review of reported cases and recommendations for genetic testing and surveillance. Genet Med. 2016;18:325–32.
Church DN, Briggs SE, Palles C, Collaborators NSECG, Kaur K, Taylor J, Tomlinson IP, et al. DNA polymerase ε and δ exonuclease domain mutations in endometrial cancer. Hum Mol Genet. 2013;22:2820–8.
The Cancer Genome Atlas Network. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73.
Song Z, Cheng G, Xu C, Wang W, Shao Y, Zhang Y. Clinicopathological characteristics of POLE mutation in patients with non-small cell lung cancer. Lung Cancer. 2018;118:57–61.
Rayner E, van Gool IC, Palles C, et al. A panoply of errors: polymerase proofreading domain mutations in cancer. Nat Rev Cancer. 2016;16:71–81.
Domingo E, Freeman-Mills L, Rayner E, et al. Somatic POLE proofreading domain mutation, immune response, and prognosis in colorectal cancer: a retrospective, pooled biomarker study. Lancet Gastroenterol Hepatol. 2016;1:207–16.
Guerra J, Pinto C, Pinto D, et al. POLE somatic mutations in advanced colorectal cancer. Cancer Med. 2017;6:2966–71.
Howitt BE, Shukla SA, Sholl LM, et al. Association of polymerase e-mutated and microsatellite-instable endometrial cancers with neoantigen load, number of tumor-infiltrating lymphocytes, and expression of PD-1 and PD-L1. JAMA Oncol. 2015;1:1319–23.
Le DT, Durham JN, Smith KN, et al. PD-1 blockade in tumors in tumors with mismatch—repair deficiency. N Engl J Med. 2015;372:2509–20.
Gong J, Wang C, Lee PP, Chu P, Fakih M. Response to PD-1 blockade in microsatellite stable metastatic colorectal cancer harboring a POLE mutations. J Natl Canc Netw. 2017;15:142–7.
Ahn SM, Ansari AA, Kim J, et al. The somatic POLE P286R mutation defines a unique subclass of colorectal cancer featuring hypermutation, representing a potential genomic biomarker for immunotherapy. Oncotarget. 2017;7:68638–49.
Connell LC, Mota JM, Braghiroli MI, Hoff PM. The rising incidence of younger patients with colorectal cancer: questions about screening, biology, and treatment. Curr Treat Options Oncol. 2017;18:23.
Schellerer VS, Merkel S, Schumann SC, et al. Despite aggressive histopathology survival is not impaired in young patients with colorectal cancer: CRC in patietns under 50 years of age. Int J Colorectal Dis. 2012;27:71–9.
Murphy CC, Singal AG. Establishing a research agenda for early-onset colorectal cancer. PLOS Med. 2018;15:e1002577.
Temko D, Van Gool IC, Rayner E, et al. Somatic POLE exonuclease domain mutation are early events in sporadic endometrial and colorectal carcinogenesis, determining driver mutational landscape, clonal neoantigen burden and immune response. J Pathol. 2018;245:283–96.
Kašubová I, Kalman M, Jašek K, et al. Stratification of patients with colorectal cancer without the recorded family history. Oncol Lett. 2019;17:3649–56.
Sobin LH, Gospodarowicz MK, Wittekind C. TNM classification of malignant tumours. 7th ed. New York: Wiley; 2009.
Edge SB, Byrd SR, Compton CC. AJCC cancer staging manual. 7th ed. New York: Springer; 2010.
Jenkins MA, Hayashi S, O’Shea AM, et al. Colon Cancer Family Registry. Pathology features in Bethesda guidelines predict colorectal cancer microsatellite instability: a population—based study. Gastroenterology. 2007;133:48–56.
Hyde A, Fontaine D, Stuckless S, et al. A histology-based model for predicting microsatellite instability in colorectal cancers. Am J Surg Pathol. 2010;34:1820–9.
Malicherova B, Burjanivova T, Grendar M, et al. Droplet digital PCR for detection of BRAF V600E mutation in formalin-fixed, paraffin-embedded melanoma tissues: a comparison with Cobas 4800, Sanger sequencing, and allele-specific PCR. Am J Transl Res. 2018;10:3773–81.
Jasek K, Buzalkova V, Minarik G, et al. Detection of mutations in the BRAF gene in patients with KIT and PDGFRA wild-type gastrointestinal stromal tunors. Virchows Arch. 2017;470:29–36.
Esteban-Jurado C, Giménez-Zaragoza D, Muñoz J, et al. POLE and POLD1 screening in 155 patients with multiple polyps and early-onset colorectal cancer. Oncotarget. 2017;8:26732–43.
Wang C, Gong J, Tu TY, Lee PP, Fakih M. Immune profiling of microsatellite instability-high and polymerase ε (POLE)- mutated metastatic colorectal tumors identifies predictors of response to anti-PD-1 therapy. J Gastrointest Oncol. 2018;9:404–15.
Grasso CS, Giannakis M, Wells DK, et al. Genetic mechanisms of immune evasion in colorectal cancer. Cancer Discov. 2018;8:730–49.
Funding
This study was supported by the Biomedical center Martin (ITMS 26220220187), which is co-financed from EU sources, by Government of Russian Federation, Grant 08-08, and by the projects of the Slovak Research and development Agency APVV Grant APVV-16-0066, and VEGA Grants 1/0199/17 and 1/0380/18.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lasabová, Z., Kalman, M., Holubeková, V. et al. Mutation analysis of POLE gene in patients with early-onset colorectal cancer revealed a rare silent variant within the endonuclease domain with potential effect on splicing. Clin Exp Med 19, 393–400 (2019). https://doi.org/10.1007/s10238-019-00558-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10238-019-00558-7