Abstract
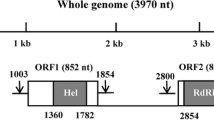
The complete genome of a novel virus from Nesidiocoris tenuis was determined by RNA-seq and rapid amplification of cDNA ends. This virus has a single-stranded RNA genome of 10633 nucleotides (nt) in length, not including the poly(A) tail, and contains two putative open reading frames (ORFs). ORF1 encodes a polypeptide of 1320 amino acids (aa) with a predicted molecular mass of 147.92 kDa and theoretical isoelectric point (pI) of 6.96. ORF2 encodes a polypeptide of 1728 aa with a predicted molecular mass of 197.09 kDa and pI of 6.73. Phylogenetic analysis with the deduced aa sequences of the conserved RNA dependent RNA polymerase domain as well as whole genome nt sequences indicated that the virus clusters with viruses classified within the genus Iflavirus, with a high bootstrap value in the maximum-likelihood and neighbor-joining trees. However, this virus has a distinct genome structure with two ORFs, iflaviruses normally having one, suggesting the virus might be a prototype of a new genus. We named the virus isolate Nesidiocoris tenuis virus 1 (NtV-1). The prevalence of NtV-1 infection in wild samples of N. tenuis was at a low level (7.32%, 6 positive in 82 samples), suggesting a possible harmful effect to its host.


Similar content being viewed by others
References
Valles SM, Chen Y, Firth AE, Guérin DMA, Hashimoto Y, Herrero S, de Miranda JR, Ryabov E, ICTV Report Consortium (2017) ICTV Virus Taxonomy Profiles: Iflaviridae. J Gen Virol 98:527–528
Geng P, Li W, de Miranda JR, Qian Z, An L, Terenius O (2017) Studies on the transmission and tissue distribution of Antheraea pernyi iflavirus in the Chinese oak silkmoth Antheraea pernyi. Virol 502:171–175
Geng P, Li W, Lin L, de Miranda JR, Emrich S, An L, Terenius O (2014) Genetic characterization of a novel Iflavirus associated with vomiting disease in the Chinese oak silkmoth Antheraea pernyi. PLoS One 9:e92107
Kalynych S, Fuzik T, Pridal A, de Miranda J, Plevka P (2017) Cryo-EM study of slow bee paralysis virus at low pH reveals iflavirus genome release mechanism. Proc Natl Acad Sci U S A 114:598–603
Kalynych S, Pridal A, Palkova L, Levdansky Y, de Miranda JR, Plevka P (2016) Virion structure of iIflavirus slow bee paralysis virus at 2.6-angstrom resolution. J Virol 90:7444–7455
Ryabov EV, Wood GR, Fannon JM, Moore JD, Bull JC, Chandler D, Mead A, Burroughs N, Evans DJ (2014) A virulent strain of deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor-mediated, or in vitro, transmission. PLoS Pathog 10:e1004230
Silva LA, Ardisson-Araujo DM, Tinoco RS, Fernandes OA, Melo FL, Ribeiro BM (2015) Complete genome sequence and structural characterization of a novel iflavirus isolated from Opsiphanes invirae (Lepidoptera: Nymphalidae). J Invertebr Pathol 130:136–140
Carrillo-Tripp J, Krueger EN, Harrison RL, Toth AL, Miller WA, Bonning BC (2014) Lymantria dispar iflavirus 1 (LdIV1), a new model to study iflaviral persistence in lepidopterans. J Gen Virol 95:2285–2296
Liu S, Chen Y, Sappington TW, Bonning BC (2017) Genome sequence of the first coleopteran iflavirus isolated from western corn rootworm. Diabrotica virgifera virgifera LeConte, Genome Announc, p 5
Murakami R, Suetsugu Y, Kobayashi T, Nakashima N (2013) The genome sequence and transmission of an iflavirus from the brown planthopper, Nilaparvata lugens. Virus Res 176:179–187
Murakami R, Suetsugu Y, Nakashima N (2014) Complete genome sequences of two iflaviruses from the brown planthopper, Nilaparvata lugens. Arch Virol 159:585–588
Yuan H, Xu P, Yang X, Graham RI, Wilson K, Wu K (2017) Characterization of a .novel member of genus Iflavirus in Helicoverpa armigera. J Invertebr Pathol 144:65–73
Arno J, Castane C, Riudavets J, Gabarra R (2010) Risk of damage to tomato crops by the generalist zoophytophagous predator Nesidiocoris tenuis (Reuter) (Hemiptera: Miridae). Bull Entomol Res 100:105–115
Nakaishi K, Arakawa R (2011) Effects of spiracle-blocking insecticides and microbial insecticides on the predator mirid bug, Nesidiocoris tenuis (reuter) (heteroptera: miridae). Pak J Biol Sci 14:991–995
Sanchez JA, Lacasa A (2008) Impact of the zoophytophagous plant bug Nesidiocoris tenuis (Heteroptera: Miridae) on tomato yield. J Econ Entomol 101:1864–1870
Xu P, Song X, Yang X, Tang Z, Ren G, Lu Y (2017) A novel single-stranded RNA virus in Nesidiocoris tenuis. Arch Virol 162:1125–1128
Wang J, Zhang J, Jiang H, Liu C, Yi F, Hu Y (2005) Nucleotide sequence and genomic organization of a newly isolated densovirus infecting Dendrolimus punctatus. J Gen Virol 86:2169–2173
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol 33:1870–1874
Cotmore SF, Agbandje-McKenna M, Chiorini JA, Mukha DV, Pintel DJ, Qiu J, Soderlund-Venermo M, Tattersall P, Tijssen P, Gatherer D, Davison AJ (2014) The family Parvoviridae. Arch Virol 159:1239–1247
Luria N, Reingold V, Lachman O, Sela N, Dombrovsky A (2016) Extended phylogenetic analysis of a new Israeli isolate of Brevicoryne brassicae virus (BrBV-IL) suggests taxonomic revision of the genus Iflavirus. Virol J 13:50
Xu P, Graham RI, Wilson K, Wu K (2017) Structure and transcription of the Helicoverpa armigera densovirus (HaDV2) genome and its expression strategy in LD652 cells. Virol J 14:23
Xu P, Liu Y, Graham RI, Wilson K, Wu K (2014) Densovirus is a mutualistic symbiont of a gGlobal crop pest (Helicoverpa armigera) and protects against a baculovirus and Bt Biopesticide. PLoS Pathog 10:e1004490
Funding
This study was funded by the National Natural Science Foundation of China (Grant No. 31401752) and the Agricultural Science and Technology Innovation Program (Grant No. ASTIP-TRIC04).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no competing interests.
Ethical approval
No permits were required for the described insect collection and experimentation.
Additional information
Handling Editor: T. K. Frey.
Nucleotide sequence accession number. The complete genome sequence of NtV-1 was submitted to GenBank with the accession number KY969634.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dong, Y., Chao, J., Liu, J. et al. Characterization of a novel RNA virus from Nesidiocoris tenuis related to members of the genus Iflavirus . Arch Virol 163, 571–574 (2018). https://doi.org/10.1007/s00705-017-3622-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3622-8