Abstract
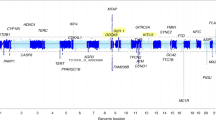
Keloids are benign dermal tumors that occur ~20 times more often in African versus Caucasian descent individuals. While most keloids occur sporadically, a genetic predisposition is supported by both familial aggregation of some keloids and the large differences in risk among populations. Yet, no well-established genetic risk factors for keloids have been identified. In this study, we conducted admixture mapping and whole-exome association using 478 African Americans (AAs) samples (122 cases, 356 controls) with exome genotyping data to identify regions where local ancestry associated with keloid risk. Logistic regression was used to evaluate associations under admixture peaks. A significant mapping peak was observed on chr15q21.2–22.3. This peak included NEDD4, a gene previously implicated in a keloid genome-wide association study (GWAS) of Japanese individuals later validated in a Chinese cohort. While we observed modest evidence for association with NEDD4, a more significant association was observed at (myosin 1E) MYO1E. A genome scan not including the 15q21-22 region also identified associations at MYO7A (rs35641839, odds ratio [OR] = 4.71, 95 % confidence interval [CI] 2.38–9.32, p = 8.34 × 10−6) at 11q13.5. The identification of SNPs in two myosin genes strongly associated with keloid formation suggests that an altered cytoskeleton contributes to the enhanced migratory and invasive properties of keloid fibroblasts. Our findings support the admixture mapping approach for the study of keloid risk, and indicate potentially common genetic elements on chr15q21.2–22.3 in causation of keloids in AAs, Japanese, and Chinese populations.



Similar content being viewed by others
References
Adeyemo A, Gerry N, Chen G, Herbert A, Doumatey A, Huang H, Zhou J, Lashley K, Chen Y, Christman M, Rotimi C (2009) A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet 5:e1000564
Anum EA, Springel EH, Shriver MD, Strauss JF III (2009) Genetic contributions to disparities in preterm birth. Pediatr Res 65:1–9
Barrett J (1973) Keloid. In: Bergsma D (ed) Birth defect compendium. Williams and Wilkens Company, Baltimore
Bond JE, Bergeron A, Thurlow P, Selim MA, Bowers EV, Kuang A, Levinson H (2011) Angiotensin-II mediates nonmuscle myosin II activation and expression and contributes to human keloid disease progression. Mol Med 17:1196–1203
Butler PD, Longaker MT, Yang GP (2008) Current progress in keloid research and treatment. J Am Coll Surg 206:731–741
Cheng J, Grassart A, Drubin DG (2012) Myosin 1E coordinates actin assembly and cargo trafficking during clathrin-mediated endocytosis. Mol Biol Cell 23:2891–2904
Chevray PM, Manson PN (2004) Keloid scars are formed by polyclonal fibroblasts. Ann Plast Surg 52:605–608
Cooper RS, Tayo B, Zhu X (2008) Genome-wide association studies: implications for multiethnic samples. Hum Mol Genet 17:R151–R155
Davis KD, Garcia M, Phillips JA III, Hedges L, Haines J, Carneal J et al (2000) Detection of a critical interval for a familial keloid locus on chromosome 14q22-q23 in an African–American pedigree. In: Presented at the 50th annual meeting of the American society of human genetics, Philadelphia, PA
Fernandez LP, Milne RL, Pita G, Floristan U, Sendagorta E, Feito M, Aviles JA, Martin-Gonzalez M, Lazaro P, Benitez J, Ribas G (2009) Pigmentation-related genes and their implication in malignant melanoma susceptibility. Exp Dermatol 18:634–642
Flores C, Ma SF, Pino-Yanes M, Wade MS, Perez-Mendez L, Kittles RA, Wang D, Papaiahgari S, Ford JG, Kumar R, Garcia JG (2012) African ancestry is associated with asthma risk in African Americans. PLoS One 7:e26807
Howie BN, Donnelly P, Marchini J (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5:e1000529
Howie B, Marchini J, Stephens M (2011) Genotype imputation with thousands of genomes. G3 1:457–470
Inouye M, Ripatti S, Kettunen J, Lyytikainen LP, Oksala N, Laurila PP, Kangas AJ, Soininen P, Savolainen MJ, Viikari J, Kahonen M, Perola M, Salomaa V, Raitakari O, Lehtimaki T, Taskinen MR, Jarvelin MR, Ala-Korpela M, Palotie A, de Bakker PI (2012) Novel loci for metabolic networks and multi-tissue expression studies reveal genes for atherosclerosis. PLoS Genet 8:e1002907
Liu XZ, Hope C, Walsh J, Newton V, Ke XM, Liang CY, Xu LR, Zhou JM, Trump D, Steel KP, Bundey S, Brown SD (1998) Mutations in the myosin VIIA gene cause a wide phenotypic spectrum, including atypical Usher syndrome. Am J Hum Genet 63:909–912
Marchini J, Howie B, Myers S, McVean G, Donnelly P (2007) A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 39:906–913
Marneros AG, Norris JE, Watanabe S, Reichenberger E, Olsen BR (2004) Genome scans provide evidence for keloid susceptibility loci on chromosomes 2q23 and 7p11. J Invest Dermatol 122:1126–1132
Mele C, Iatropoulos P, Donadelli R, Calabria A, Maranta R, Cassis P, Buelli S, Tomasoni S, Piras R, Krendel M, Bettoni S, Morigi M, Delledonne M, Pecoraro C, Abbate I, Capobianchi MR, Hildebrandt F, Otto E, Schaefer F, Macciardi F, Ozaltin F, Emre S, Ibsirlioglu T, Benigni A, Remuzzi G, Noris M (2011) MYO1E mutations and childhood familial focal segmental glomerulosclerosis. N Engl J Med 365:295–306
Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, von ME, Farrall M, Lathrop M, Cookson WO (2010) A large-scale, consortium-based genomewide association study of asthma. N Engl J Med 363:1211–1221
Moulton-Levy P, Jackson CE, Levy HG, Fialkow PJ (1984) Multiple cell origin of traumatically induced keloids. J Am Acad Dermatol 10:986–988
Nakashima M, Chung S, Takahashi A, Kamatani N, Kawaguchi T, Tsunoda T, Hosono N, Kubo M, Nakamura Y, Zembutsu H (2010) A genome-wide association study identifies four susceptibility loci for keloid in the Japanese population. Nat Genet 42:768–771
Niessen FB, Spauwen PH, Schalkwijk J, Kon M (1999) On the nature of hypertrophic scars and keloids: a review. Plast Reconstr Surg 104:1435–1458
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909
Pulley J, Clayton E, Bernard GR, Roden DM, Masys DR (2010) Principles of human subjects protections applied in an opt-out, de-identified biobank. Clin Transl Sci 3:42–48
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575
Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, Boehnke M (2010) Genome-wide association studies in diverse populations. Nat Rev Genet 11:356–366
Roux AF, Faugere V, Le GS, Pallares-Ruiz N, Vielle A, Chambert S, Marlin S, Hamel C, Gilbert B, Malcolm S, Claustres M (2006) Survey of the frequency of USH1 gene mutations in a cohort of Usher patients shows the importance of cadherin 23 and protocadherin 15 genes and establishes a detection rate of above 90 %. J Med Genet 43:763–768
Russell JD, Russell SB, Trupin KM (1978) Differential effects of hydrocortisone on both growth and collagen metabolism of human fibroblasts from normal and keloid tissue. J Cell Physiol 97:221–229
Russell SB, Russell JD, Trupin KM, Gayden AE, Opalenik SR, Nanney LB, Broquist AH, Raju L, Williams SM (2010) Epigenetically altered wound healing in keloid fibroblasts. J Invest Dermatol 130:2489–2496
Saed GM, Ladin D, Olson J, Han X, Hou Z, Fivenson D (1998) Analysis of p53 gene mutations in keloids using polymerase chain reaction-based single-strand conformational polymorphism and DNA sequencing. Arch Dermatol 134:963–967
Sankararaman S, Sridhar S, Kimmel G, Halperin E (2008) Estimating local ancestry in admixed populations. Am J Hum Genet 82:290–303
Smith JC, Boone BE, Opalenik SR, Williams SM, Russell SB (2008) Gene profiling of keloid fibroblasts shows altered expression in multiple fibrosis-associated pathways. J Invest Dermatol 128:1298–1310
Trupin JS, Williams J, Hammons J, Russell J (1977) Multicellular origin of keloids. Birth defects: Fifth International Conference. Elsevier, Montreal, pp 121
Winkler CA, Nelson GW, Smith MW (2010) Admixture mapping comes of age. Annu Rev Genomics Hum Genet 11:65–89
Zhu X, Luke A, Cooper RS, Quertermous T, Hanis C, Mosley T, Gu CC, Tang H, Rao DC, Risch N, Weder A (2005) Admixture mapping for hypertension loci with genome-scan markers. Nat Genet 37:177–181
Zhu X, Young JH, Fox E, Keating BJ, Franceschini N, Kang S, Tayo B, Adeyemo A, Sun YV, Li Y, Morrison A, Newton-Cheh C, Liu K, Ganesh SK, Kutlar A, Vasan RS, Dreisbach A, Wyatt S, Polak J, Palmas W, Musani S, Taylor H, Fabsitz R, Townsend RR, Dries D, Glessner J, Chiang CW, Mosley T, Kardia S, Curb D, Hirschhorn JN, Rotimi C, Reiner A, Eaton C, Rotter JI, Cooper RS, Redline S, Chakravarti A, Levy D (2011) Combined admixture mapping and association analysis identifies a novel blood pressure genetic locus on 5p13: contributions from the CARe consortium. Hum Mol Genet 20:2285–2295
Zhu F, Wu B, Li P, Wang J, Tang H, Liu Y, Zuo X, Cheng H, Ding Y, Wang W, Zhai Y, Qian F, Wang W, Yuan X, Wang J, Ha W, Hou J, Zhou F, Wang Y, Gao J, Sheng Y, Sun L, Liu J, Yang S, Zhang X (2013) Association study confirmed susceptibility loci with keloid in the Chinese Han population. PLoS One 8:e62377
Acknowledgments
This work was supported by NIH Grants F33AR052241 (SBR), P50DE10595 (SBR), R03AR048938 (SMW), P30AR041943 (SMW), P20GM103534 (SMW), pilot Grant from the Center for Human Genetics Research at VU (SBR, DRVE and TLE), 1UL1RR024975 (SBR), 5KL2TR000446-08 (TLE), 5K12HD04383-12 (DRVE) and by resources of Vanderbilt University School of Medicine and University of North Carolina Functional Genomics Core. The Project described was also supported by CTSA award No. UL1TR000445 from the National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent official views of the National Center for Advancing Translational Sciences or the NIH.
Conflict of interest
None declared.
Author information
Authors and Affiliations
Corresponding authors
Additional information
D. R. Velez Edwards and K. S. Tsosie are co-first author equal contribution.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. 1. Quality control flow chart
Supplemental Fig. 2. Quantile–quantile (Q–Q) plot of LAMP analyses. Expected −log10 p values are on the x-axis and the observed −log10 p values are on the y-axis.
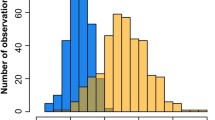
Supplemental Fig. 3. Multidimensional scaling plots of keloid cases and controls with HapMap populations. MDS axes for AAs were plotted with MDS1 on the y-axis and MDS2 on the x-axis. Values were color coded according to self-reported race (AA light blue circles keloid cases, AA dark blue circles controls, AA HapMap reference population (ASW) black circles, European American HapMap reference population (CEU) red circles, and African HapMap reference population (YRI) green circles).
Supplemental Fig. 4. Summary of strongest admixture mapping peaks across all chromosomes. X-axis indicates genomic position along the chromosome. Y-axis indicates −log10 (p value) from logistic regression (adjusted for sex and top 10 PCs) of admixture mapping values generated from LAMP. Dotted line indicates threshold of significance, as determined by negative log value of the family-wise error rate (FWER), calculated from n = 1,000 permutations in multiple testing (top line), the bottom line is an arbitrary threshold that is one log down from the FWER.
Rights and permissions
About this article
Cite this article
Velez Edwards, D.R., Tsosie, K.S., Williams, S.M. et al. Admixture mapping identifies a locus at 15q21.2–22.3 associated with keloid formation in African Americans. Hum Genet 133, 1513–1523 (2014). https://doi.org/10.1007/s00439-014-1490-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-014-1490-9