Abstract
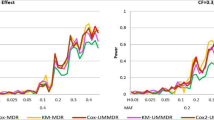
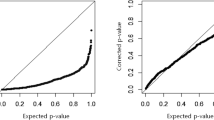
The widespread use of high-throughput methods of single nucleotide polymorphism (SNP) genotyping has created a number of computational and statistical challenges. The problem of identifying SNP–SNP interactions in case–control studies has been studied extensively, and a number of new techniques have been developed. Little progress has been made, however, in the analysis of SNP–SNP interactions in relation to time-to-event data, such as patient survival time or time to cancer relapse. We present an extension of the two class multifactor dimensionality reduction (MDR) algorithm that enables detection and characterization of epistatic SNP–SNP interactions in the context of survival analysis. The proposed Survival MDR (Surv-MDR) method handles survival data by modifying MDR’s constructive induction algorithm to use the log-rank test. Surv-MDR replaces balanced accuracy with log-rank test statistics as the score to determine the best models. We simulated datasets with a survival outcome related to two loci in the absence of any marginal effects. We compared Surv-MDR with Cox-regression for their ability to identify the true predictive loci in these simulated data. We also used this simulation to construct the empirical distribution of Surv-MDR’s testing score. We then applied Surv-MDR to genetic data from a population-based epidemiologic study to find prognostic markers of survival time following a bladder cancer diagnosis. We identified several two-loci SNP combinations that have strong associations with patients’ survival outcome. Surv-MDR is capable of detecting interaction models with weak main effects. These epistatic models tend to be dropped by traditional Cox regression approaches to evaluating interactions. With improved efficiency to handle genome wide datasets, Surv-MDR will play an important role in a research strategy that embraces the complexity of the genotype–phenotype mapping relationship since epistatic interactions are an important component of the genetic basis of disease.



Similar content being viewed by others
References
Andrew AS, Gui J, Sanderson AC, Mason RA, Morlock EV, Schned AR, Kelsey KT, Marsit CJ, Moore JH, Karagas MR (2009) Bladder cancer SNP panel predicts susceptibility and survival. Hum Genet 125:527–539
Hahn LW, Moore JH (2004) Ideal discrimination of discrete clinical endpoints using multilocus genotypes. In Silico Biol 4:183–194
Hahn LW, Ritchie MD, Moore JH (2003) Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics 19:376–382
He H, Oetting WS, Brott MJ, Basu S (2009) Power of multifactor dimensionality reduction and penalized logistic regression for detecting gene-gene interaction in a case-control study. BMC Med Genet 10:127
Huang J, Lin A, Narasimhan B, Quertermous T, Hsiung CA, Ho LT, Grove JS, Olivier M, Ranade K, Risch NJ, Olshen RA (2004) Tree-structured supervised learning and the genetics of hypertension. PNAS 101:10529–10534
Kamal NS, Soria JC, Mendiboure J, Planchard D, Olaussen KA, Rousseau V, Popper H, Pirker R, Bertrand P, Dunant A, Le Chevalier T, Filipits M et al (2010) MutS homologue 2 and the long-term benefit of adjuvant chemotherapy in lung cancer. Clin Cancer Res 16:1206–1215
Lou XY, Chen GB, Yan L, Ma JZ, Zhu J, Elston RC, Li MD (2007) A generalized combinatorial approach for detecting gene by gene and gene by environment interactions with application to nicotine dependence. Am J Hum Genet 80:1125–1137
Michalski RS (1983) A theory and methodology of inductive learning. Artif Intell 20:111–161
Moore JH (2004) Computational analysis of gene-gene interactions in common human diseases using multifactor dimensionality reduction. Expert Rev Mol Diagn 4:795–803
Moore JH (2007) Genome-wide analysis of epistasis using multifactor dimensionality reduction: feature selection and construction in the domain of human genetics. In: Zhu X, Davidson I (eds) Knowledge discovery and data mining: challenges and realities with real world data. IGI Press, Hershey, pp 17–30
Moore JH, Williams SM (2009) Epistasis and its implications for personal genetics. Am J Hum Genet 85:309–320
Moore JH, Gilbert JC, Tsai CT, Chiang FT, Holden W, Barney N, White BC (2006) A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J Theor Biol 241:252–261
Moore JH, Asselbergs FW, Williams SM (2010) Bioinformatics challenges for genome-wide association studies. Bioinformatics 26:445–455
Park M, Hastie T (2008) Penalized logistic regression for detecting gene interactions. Biostatistics 9:30–50
Qiu C, Yu M, Shan L, Snyderwine EG (2003) Allelic imbalance and altered expression of genes in chromosome 2q11–2q16 from rat mammary gland carcinomas induced by 2-amino-1-methyl-6-phenylimidazo pyridine. Oncogene 22:1253–1260
Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH (2001) Multifactor dimensionality reduction reveals high-order interactions among estrogen metabolism genes in sporadic breast cancer. Am J Hum Genet 69:138–147
Ritchie MD, Hahn LW, Moore JH (2003) Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genet Epidemiol 24:150–157
Seki M, Otsuki M, Ishii Y, Tada S, Enomoto T (2008) RecQ family helicases in genome stability: lessons from gene disruption studies in DT40 cells. Cell Cycle 7:2472–2478
Velez DR, White BC, Motsinger AA, Bush WS, Ritchie MD, Williams SM, Moore JH (2007) A balanced accuracy metric for epistasis modeling in imbalanced datasets using multifactor dimensionality reduction. Genet Epidemiol 31:306–315
Yan L, Verbel D, Saidi O (2004) Predicting prostate cancer recurrence via maximizing the concordance index. In: Proceedings of the 10th ACM SIGKDD international conference on knowledge discovery and data mining, pp 479–485
Acknowledgments
This work was funded by grant #IRG-82-003-22 from the American Cancer Society and NIH grants LM009012, LM010098, AI59694, CA078609, CA121382, CA102327, CA57494 and ES007373.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gui, J., Moore, J.H., Kelsey, K.T. et al. A novel survival multifactor dimensionality reduction method for detecting gene–gene interactions with application to bladder cancer prognosis. Hum Genet 129, 101–110 (2011). https://doi.org/10.1007/s00439-010-0905-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-010-0905-5