Abstract
Messenger RNA (mRNA) markers have been extensively investigated for the identification of forensically relevant body fluids and tissues based on their expression profiles among cell types. As products of the backsplicing of pre-mRNAs, circular RNAs (circRNAs) share exonic sequences with their linear counterparts. The inclusion of circRNAs in mRNA profiling is shown to facilitate the detection of biomarkers in the identification of body fluids. In this study, we identified the expression of circRNAs of 14 out of 45 biomarkers from five body fluid types using outward-facing primer sets and revealed the ratio of circular to total transcripts of biomarkers by RNase R treatment. Furthermore, our results of qPCR analysis show that the inclusion of circRNAs in the detection of biomarkers, including HBA and ALAS2 for blood; MMP7 and MMP10 for menstrual blood; HTN3 for saliva; SPINK5, SERPINB3, ESR1, and CYP2B7P1 for vaginal secretions; TGM4, KLK3, and PRM2 for semen; and SLC22A6 and MIOX for urine, does not impair the specificity of these biomarkers. Additionally, a high copy number of targets from linear transcripts could be employed to increase the detection sensitivity of TGM4 and KLK3 with a low expression level of circRNAs in urine samples. Altogether, these results will help with the development of robust multiplex assays for body fluid identification.




Similar content being viewed by others
References
Roeder AD, Haas C (2013) MRNA profiling using a minimum of five mRNA markers per body fluid and a novel scoring method for body fluid identification. Int J Legal Med 127:707–721. https://doi.org/10.1007/s00414-012-0794-3
Juusola J, Ballantyne J (2005) Multiplex mRNA profiling for the identification of body fluids. Forensic Sci Int 152:1–12. https://doi.org/10.1016/j.forsciint.2005.02.020
Hanson EK, Ballantyne J (2013) Highly specific mRNA biomarkers for the identification of vaginal secretions in sexual assault investigations. Sci Justice 53:14–22. https://doi.org/10.1016/j.scijus.2012.03.007
Zubakov D, Hanekamp E, Kokshoorn M, van IJcken W, Kayser M (2008) Stable RNA markers for identification of blood and saliva stains revealed from whole genome expression analysis of time-wise degraded samples. Int J Legal Med 122:135–142. https://doi.org/10.1007/s00414-007-0182-6
Lindenbergh A, Maaskant P, Sijen T (2013) Implementation of RNA profiling in forensic casework. Forensic Sci Int Genet 7:159–166. https://doi.org/10.1016/j.fsigen.2012.09.003
Lindenbergh A, De Pagter M, Ramdayal G et al (2012) A multiplex (m)RNA-profiling system for the forensic identification of body fluids and contact traces. Forensic Sci Int Genet 6:565–577. https://doi.org/10.1016/j.fsigen.2012.01.009
Visser M, Zubakov D, Ballantyne KN, Kayser M (2011) MRNA-based skin identification for forensic applications. Int J Legal Med 125:253–263. https://doi.org/10.1007/s00414-010-0545-2
Park JL, Park SM, Kwon OH, Lee HC, Kim JY, Seok HH, Lee WS, Lee SH, Kim YS, Woo KM, Kim SY (2014) Microarray screening and qRT-PCR evaluation of microRNA markers for forensic body fluid identification. Electrophoresis 35:3062–3068. https://doi.org/10.1002/elps.201400075
Sauer E, Reinke AK, Courts C (2016) Differentiation of five body fluids from forensic samples by expression analysis of four microRNAs using quantitative PCR. Forensic Sci Int Genet 22:89–99. https://doi.org/10.1016/j.fsigen.2016.01.018
Wang Z, Zhang J, Luo H, Ye Y, Yan J, Hou Y (2013) Screening and confirmation of microRNA markers for forensic body fluid identification. Forensic Sci Int Genet 7:116–123. https://doi.org/10.1016/j.fsigen.2012.07.006
Sauer E, Babion I, Madea B, Courts C (2014) An evidence based strategy for normalization of quantitative PCR data from miRNA expression analysis in forensically relevant body fluids. Forensic Sci Int Genet 13:217–223. https://doi.org/10.1016/j.fsigen.2014.08.005
Choi A, Shin KJ, Yang WI, Lee HY (2014) Body fluid identification by integrated analysis of DNA methylation and body fluid-specific microbial DNA. Int J Legal Med 128:33–41. https://doi.org/10.1007/s00414-013-0918-4
Kader F, Ghai M (2015) DNA methylation and application in forensic sciences. Forensic Sci Int 249:255–265. https://doi.org/10.1016/j.forsciint.2015.01.037
Vidaki A, Giangasparo F, Syndercombe Court D (2016) Discovery of potential DNA methylation markers for forensic tissue identification using bisulphite pyrosequencing. Electrophoresis 37:2767–2779. https://doi.org/10.1002/elps.201600261
Lee HY, Jung SE, Lee EH, Yang WI, Shin KJ (2016) DNA methylation profiling for a confirmatory test for blood, saliva, semen, vaginal fluid and menstrual blood. Forensic Sci Int Genet 24:75–82. https://doi.org/10.1016/j.fsigen.2016.06.007
An JH, Choi A, Shin KJ, Yang WI, Lee HY (2013) DNA methylation-specific multiplex assays for body fluid identification. Int J Legal Med 127:35–43. https://doi.org/10.1007/s00414-012-0719-1
Hanssen EN, Avershina E, Rudi K, Gill P, Snipen L (2017) Body fluid prediction from microbial patterns for forensic application. Forensic Sci Int Genet 30:10–17. https://doi.org/10.1016/j.fsigen.2017.05.009
Ingold S, Dørum G, Hanson E, Berti A, Branicki W, Brito P, Elsmore P, Gettings KB, Giangasparo F, Gross TE, Hansen S, Hanssen EN, Kampmann ML, Kayser M, Laurent FX, Morling N, Mosquera-Miguel A, Parson W, Phillips C, Porto MJ, Pośpiech E, Roeder AD, Schneider PM, Schulze Johann K, Steffen CR, Syndercombe-Court D, Trautmann M, van den Berge M, van der Gaag KJ, Vannier J, Verdoliva V, Vidaki A, Xavier C, Ballantyne J, Haas C (2018) Body fluid identification using a targeted mRNA massively parallel sequencing approach—results of a EUROFORGEN/EDNAP collaborative exercise. Forensic Sci Int Genet 34:105–115. https://doi.org/10.1016/j.fsigen.2018.01.002
Haas C, Hanson E, Anjos MJ, Ballantyne KN, Banemann R, Bhoelai B, Borges E, Carvalho M, Courts C, de Cock G, Drobnic K, Dötsch M, Fleming R, Franchi C, Gomes I, Hadzic G, Harbison SA, Harteveld J, Hjort B, Hollard C, Hoff-Olsen P, Hüls C, Keyser C, Maroñas O, McCallum N, Moore D, Morling N, Niederstätter H, Noël F, Parson W, Phillips C, Popielarz C, Roeder AD, Salvaderi L, Sauer E, Schneider PM, Shanthan G, Court DS, Turanská M, van Oorschot RAH, Vennemann M, Vidaki A, Zatkalíková L, Ballantyne J (2014) RNA/DNA co-analysis from human menstrual blood and vaginal secretion stains: results of a fourth and fifth collaborative EDNAP exercise. Forensic Sci Int Genet 8:203–212. https://doi.org/10.1016/j.fsigen.2013.09.009
Haas C, Hanson E, Banemann R, Bento AM, Berti A, Carracedo Á, Courts C, Cock GD, Drobnic K, Fleming R, Franchi C, Gomes I, Hadzic G, Harbison SA, Hjort B, Hollard C, Hoff-Olsen P, Keyser C, Kondili A, Maroñas O, McCallum N, Miniati P, Morling N, Niederstätter H, Noël F, Parson W, Porto MJ, Roeder AD, Sauer E, Schneider PM, Shanthan G, Sijen T, Syndercombe Court D, Turanská M, van den Berge M, Vennemann M, Vidaki A, Zatkalíková L, Ballantyne J (2015) RNA/DNA co-analysis from human skin and contact traces—results of a sixth collaborative EDNAP exercise. Forensic Sci Int Genet 16:139–147. https://doi.org/10.1016/j.fsigen.2015.01.002
Haas C, Hanson E, Anjos MJ, Banemann R, Berti A, Borges E, Carracedo A, Carvalho M, Courts C, de Cock G, Dötsch M, Flynn S, Gomes I, Hollard C, Hjort B, Hoff-Olsen P, Hríbiková K, Lindenbergh A, Ludes B, Maroñas O, McCallum N, Moore D, Morling N, Niederstätter H, Noel F, Parson W, Popielarz C, Rapone C, Roeder AD, Ruiz Y, Sauer E, Schneider PM, Sijen T, Court DS, Sviežená B, Turanská M, Vidaki A, Zatkalíková L, Ballantyne J (2013) RNA/DNA co-analysis from human saliva and semen stains—results of a third collaborative EDNAP exercise. Forensic Sci Int Genet 7:230–239. https://doi.org/10.1016/j.fsigen.2012.10.011
Cossu C, Germann U, Kratzer A, Bär W, Haas C (2009) How specific are the vaginal secretion mRNA-markers HBD1 and MUC4? Forensic Sci Int Genet Suppl Ser 2:536–537. https://doi.org/10.1016/j.fsigss.2009.08.063
Richard MLL, Harper KA, Craig RL, Onorato AJ, Robertson JM, Donfack J (2012) Evaluation of mRNA marker specificity for the identification of five human body fluids by capillary electrophoresis. Forensic Sci Int Genet 6:452–460. https://doi.org/10.1016/j.fsigen.2011.09.007
Sirker M, Schneider PM, Gomes I (2016) A 17-month time course study of human RNA and DNA degradation in body fluids under dry and humid environmental conditions. Int J Legal Med 130:1431–1438. https://doi.org/10.1007/s00414-016-1373-9
Setzer M, Juusola J, Ballantyne J (2008) Recovery and stability of RNA in vaginal swabs and blood, semen, and saliva stains. J Forensic Sci 53:296–305. https://doi.org/10.1111/j.1556-4029.2007.00652.x
Haas C, Hanson E, Kratzer A, Bär W, Ballantyne J (2011) Selection of highly specific and sensitive mRNA biomarkers for the identification of blood. Forensic Sci Int Genet 5:449–458. https://doi.org/10.1016/j.fsigen.2010.09.006
Zhang Y, Liu B, Shao C, Xu H, Xue A, Zhao Z, Shen Y, Tang Q, Xie J (2018) Evaluation of the inclusion of circular RNAs in mRNA profiling in forensic body fluid identification. Int J Legal Med 132:43–52. https://doi.org/10.1007/s00414-017-1690-7
Song F, Luo H, Xie M, Zhu H, Hou Y (2017) Microarray expression profile of circular RNAs in human body fluids. Forensic Sci Int Genet Suppl Ser 6:e55–e56. https://doi.org/10.1016/j.fsigss.2017.09.005
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338. https://doi.org/10.1038/nature11928
Xie F, Xiao P, Chen D, Xu L, Zhang B (2012) miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol 80:75–84. https://doi.org/10.1007/s11103-012-9885-2
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S (2014) CircRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56:55–66. https://doi.org/10.1016/j.molcel.2014.08.019
Hochmeister MN, Budowle B, Rudin O, Gehrig C, Borer U, Thali M, Dirnhofer R (1999) Evaluation of prostate-specific antigen (PSA) membrane test assays for the forensic identification of seminal fluid. J Forensic Sci 44:1057–1060. https://doi.org/10.1520/JFS12042J
Xu Y, Xie J, Cao Y, Zhou H, Ping Y, Chen L, Gu L, Hu W, Bi G, Ge J, Chen X, Zhao Z (2014) Development of highly sensitive and specific mRNA multiplex system (XCYR1) for forensic human body fluids and tissues identification. PLoS One 9:7–13. https://doi.org/10.1371/journal.pone.0100123
Xia S, Feng J, Lei L, Hu J, Xia L, Wang J, Xiang Y, Liu L, Zhong S, Han L, He C (2017) Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform 18:984–992. https://doi.org/10.1093/bib/bbw081
Liu YC, Li JR, Sun CH, Andrews E, Chao RF, Lin FM, Weng SL, Hsu SD, Huang CC, Cheng C, Liu CC, Huang HD (2016) CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res 44:D209–D215. https://doi.org/10.1093/nar/gkv940
Glazar P, Papavasileiou P, Rajewsky N, et al (2014) circBase : a database for circular RNAs circBase : a database for circular RNAs. 0–5. https://doi.org/10.1261/rna.043687.113.overview
Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, Chen LL, Yang L (2016) Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res 26:1277–1287. https://doi.org/10.1101/gr.202895.115
Xu T, Wu J, Han P, Zhao Z, Song X (2017) Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genomics 18(Suppl 6):680. https://doi.org/10.1186/s12864-017-4029-3
Memczak S, Papavasileiou P, Peters O, Rajewsky N (2015) Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One 10:1–13. https://doi.org/10.1371/journal.pone.0141214
Goffin F, Munaut C, Frankenne F, Perrier d’Hauterive S, Béliard A, Fridman V, Nervo P, Colige A, Foidart JM (2003) Expression pattern of metalloproteinases and tissue inhibitors of matrix-metalloproteinases in cycling human endometrium. Biol Reprod 69:976–984. https://doi.org/10.1095/biolreprod.103.015933
Mannello F, Luchetti F, Falcieri E, Papa S (2005) Multiple roles of matrix metalloproteinases during. Apoptosis 10:19–24. https://doi.org/10.1007/s10495-005-6058-7
Verma RP, Hansch C (2007) Matrix metalloproteinases (MMPs): chemical-biological functions and (Q)SARs. Bioorganic Med Chem 15:2223–2268. https://doi.org/10.1016/j.bmc.2007.01.011
Miranda KC, Bond DT, McKee M, Skog J, Păunescu TG, da Silva N, Brown D, Russo LM (2010) Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int 78:191–199. https://doi.org/10.1038/ki.2010.106
Pampalakis G, Scorilas A, Sotiropoulou G (2008) Novel splice variants of prostate-specific antigen and applications in diagnosis of prostate cancer. Clin Biochem 41:591–597. https://doi.org/10.1016/j.clinbiochem.2007.12.022
Cho S-Y, Choi K, Jeon J-H, Kim CW, Shin DM, Lee JB, Lee SE, Kim CS, Park JS, Jeong EM, Jang GY, Song KY, Kim IG (2010) Differential alternative splicing of human transglutaminase 4 in benign prostate hyperplasia and prostate cancer. Exp Mol Med 42:310–318. https://doi.org/10.3858/emm.2010.42.4.031
Del Prete MJ, Robles MS, Guáo A et al (2002) Degradation of cellular mRNA is a general early apoptosis-induced event. FASEB J 16:2003–2005. https://doi.org/10.1096/fj.02-0392fje
Funding
This work was supported by National Natural Science Foundation of China (81571853 and 81701866).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Figure S1
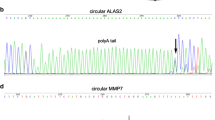
The junction sequences of head-to-tail circRNAs. Results from Sanger sequencing and the arrow indicates the junction sites of head-to-tail products. The locations of circular RNAs in the genome are indicated above each sequence. (PNG 1758 kb)
Figure S2
Target regions of genes for PCR amplification. (A) HBA and ALAS2 for blood. (B) MMP7 and MMP10 for menstrual blood. (C) HTN3 for saliva. (D) ESR1, SPINK5, SERPINB3 and CYP2B7P1 for vaginal secretions. (E) PRM2, TGM4, and KLK3 for semen. (F) MIOX and SLC22A6 for urine. The curved line indicates the downstream 3′ end of an exon is covalently linked with the upstream 5′ end of an exon, which results in the formation of a circular transcript. The regions covered by the horizontal lines represent the position of the amplicons. The blue horizontal lines indicate regions amplified by previously reported primer sets and the red horizontal lines indicate regions amplified by primer sets in this study. Asterisk and triangle indicate regions amplified by L-primers and LC-primers, respectively, in this study. (PNG 219 kb)
Figure S3
The evaluation of expression stability of candidate reference genes. The Cq values of β-actin, β2M, GAPDH and 18S rRNA for samples (n = 4) from each body fluid were used as input data in RefFinder to determine the most stable reference gene by evaluating their stability to generate scores for all candidate genes. Lower score represented more stable genes. (PNG 359 kb)
Figure S4
Dissociation curves from the real-time PCR assay using L-primers (left) and LC-primers (right) of TGM4 (A) and KLK3 (B). The amplification efficiencies were determined using L-primers of TGM4 and KLK3 as well as using LC-primers of TGM4 and KLK3 in qPCR. The cDNA from total RNA of semen was subjected to 10-fold serial dilutions. The standard curves were linear over four to five orders of magnitude with an R2 value of more than 0.98. The slope of the linear equation was used to calculate the efficiency with the following equation: E = 10–1/slope- 1. Each point represents the mean of three replicates. (PNG 362 kb)
Table S1
(DOCX 15 kb)
Table S2
(DOCX 17 kb)
Table S3
(DOCX 17 kb)
Table S4
(DOCX 24 kb)
Table S5
(DOCX 17 kb)
Rights and permissions
About this article
Cite this article
Liu, B., Song, F., Yang, Q. et al. Characterization of tissue-specific biomarkers with the expression of circRNAs in forensically relevant body fluids. Int J Legal Med 133, 1321–1331 (2019). https://doi.org/10.1007/s00414-019-02027-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-019-02027-y