Abstract
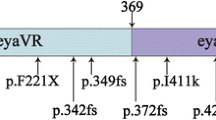
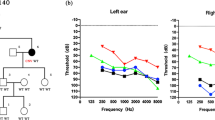
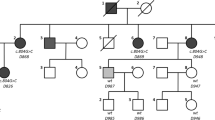
The EYA4 gene encodes a 640-amino-acid protein that serves as a transcription factor. This protein contains a highly conserved Eya domain (eya-HR) and a variable domain (eya-VR). Mutations of this gene are known to cause postlingual and progressive sensorineural hearing loss, either as non-syndromic (DFNA10) or syndromic hearing loss, depending on the location of truncation of the mutant protein. Since our previous report, we have recruited 14 families segregating autosomal dominant moderate SNHL. A thorough medical history and physical examination including evaluation of heart problems ruled out any syndromic features in these families. Screening of EYA4 was performed by targeted exome sequencing of 134 known deafness genes (TES-134) from the probands. After basic filtering of the variants, we identified one proband who carried a novel truncation mutation, c.1194delT (p.Met401TrpfsX3) of EYA4, making the frequency of DFNA10 to be 7.14 % (1/14) in Koreans. The variant co-segregated perfectly with a slightly down-sloping, moderate degree of SNHL in the family (SH117), and was not detected in any of the 592 normal control chromosomes. This variant is likely to generate protein products that are truncated just downstream of the eya-VR domain. None of the three affected family members showed any syndromic features, including cardiac problems, which was compatible with a previous genotype–phenotype correlation. The identification of a novel EYA4 truncation mutation associated with DFNA10, rather than syndromic hearing loss, supports a previously reported genotype–phenotype correlation in this gene. Considering its detection rate, EYA4 mutations should be suspected in hereditary moderate hearing loss with a corresponding audiologic configuration, and a cardiac examination should be included in the initial evaluation.




Similar content being viewed by others
References
Tekin M, Arnos KS, Pandya A (2001) Advances in hereditary deafness. Lancet 358(9287):1082–1090. doi:10.1016/S0140-6736(01)06186-4
Morton NE (1991) Genetic epidemiology of hearing impairment. Ann N Y Acad Sci 630:16–31
Petit C (1996) Genes responsible for human hereditary deafness: symphony of a thousand. Nat Genet 14(4):385–391. doi:10.1038/ng1296-385
Van Camp G, Smith RJ. Hereditary Hearing Loss Homepage. http://hereditaryhearingloss.org/. Accessed 1 May 2015
Hildebrand MS, DeLuca AP, Taylor KR, Hoskinson DP, Hur IA, Tack D, McMordie SJ, Huygen PL, Casavant TL, Smith RJ (2009) A contemporary review of AudioGene audioprofiling: a machine-based candidate gene prediction tool for autosomal dominant nonsyndromic hearing loss. Laryngoscope 119(11):2211–2215. doi:10.1002/lary.20664
Choi BY, Park G, Gim J, Kim AR, Kim BJ, Kim HS, Park JH, Park T, Oh SH, Han KH, Park WY (2013) Diagnostic application of targeted resequencing for familial nonsyndromic hearing loss. PLoS One 8(8):e68692. doi:10.1371/journal.pone.0068692
Hentze MW, Kulozik AE (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96(3):307–310
Zimmerman JE, Bui QT, Steingrimsson E, Nagle DL, Fu W, Genin A, Spinner NB, Copeland NG, Jenkins NA, Bucan M, Bonini NM (1997) Cloning and characterization of two vertebrate homologs of the Drosophila eyes absent gene. Genome Res 7(2):128–141
Wayne S, Robertson NG, DeClau F, Chen N, Verhoeven K, Prasad S, Tranebjarg L, Morton CC, Ryan AF, Van Camp G, Smith RJ (2001) Mutations in the transcriptional activator EYA4 cause late-onset deafness at the DFNA10 locus. Hum Mol Genet 10(3):195–200
Pfister M, Toth T, Thiele H, Haack B, Blin N, Zenner HP, Sziklai I, Nurnberg P, Kupka S (2002) A 4-bp insertion in the eya-homologous region (eyaHR) of EYA4 causes hearing impairment in a Hungarian family linked to DFNA10. Mol Med 8(10):607–611
Schonberger J, Levy H, Grunig E, Sangwatanaroj S, Fatkin D, MacRae C, Stacker H, Halpin C, Eavey R, Philbin EF, Katus H, Seidman JG, Seidman CE (2000) Dilated cardiomyopathy and sensorineural hearing loss: a heritable syndrome that maps to 6q23-24. Circulation 101(15):1812–1818
Schonberger J, Wang L, Shin JT, Kim SD, Depreux FF, Zhu H, Zon L, Pizard A, Kim JB, Macrae CA, Mungall AJ, Seidman JG, Seidman CE (2005) Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat Genet 37(4):418–422. doi:10.1038/ng1527
Makishima T, Madeo AC, Brewer CC, Zalewski CK, Butman JA, Sachdev V, Arai AE, Holbrook BM, Rosing DR, Griffith AJ (2007) Nonsyndromic hearing loss DFNA10 and a novel mutation of EYA4: evidence for correlation of normal cardiac phenotype with truncating mutations of the Eya domain. Am J Med Gen 143A(14):1592–1598. doi:10.1002/ajmg.a.31793
Hildebrand MS, Coman D, Yang T, Gardner RJ, Rose E, Smith RJ, Bahlo M, Dahl HH (2007) A novel splice site mutation in EYA4 causes DFNA10 hearing loss. Am J Med Gen 143A(14):1599–1604. doi:10.1002/ajmg.a.31860
Baek JI, Oh SK, Kim DB, Choi SY, Kim UK, Lee KY, Lee SH (2012) Targeted massive parallel sequencing: the effective detection of novel causative mutations associated with hearing loss in small families. Orphanet J Rare Dis 7:60. doi:10.1186/1750-1172-7-60
Conflict of interest
None of the authors has any conflict of interest, financial or otherwise.
Author information
Authors and Affiliations
Corresponding author
Additional information
H. S. Choi and A. R. Kim contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Choi, H.S., Kim, A.R., Kim, S.H. et al. Identification of a novel truncation mutation of EYA4 in moderate degree hearing loss by targeted exome sequencing. Eur Arch Otorhinolaryngol 273, 1123–1129 (2016). https://doi.org/10.1007/s00405-015-3661-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00405-015-3661-2