Abstract
Objectives
To explore the feasibility and effectiveness of machine learning (ML) based on multiparametric magnetic resonance imaging (mp-MRI) features extracted from transfer learning combined with clinical parameters to differentiate uterine sarcomas from atypical leiomyomas (ALMs).
Methods
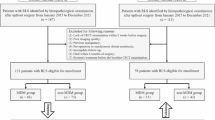
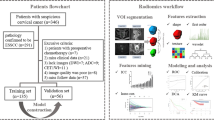
The data of 86 uterine sarcomas between July 2011 and December 2019 and 86 ALMs between June 2013 and June 2017 were retrospectively reviewed. We extracted deep-learning features and radiomics features from T2-weighted imaging (T2WI) and diffusion weighted imaging (DWI). The two feature extraction methods, transfer learning and radiomics, were compared. Random forest was adopted as the classifier. T2WI features, DWI features, combined T2WI and DWI (mp-MRI) features, and combined clinical parameters and mp-MRI features were applied to establish T2, DWI, T2-DWI, and complex multiparameter (mp) models, respectively. Predictive performance was assessed with the area under the receiver operating characteristic curve (AUC).
Results
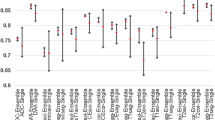
In the test set, the T2, DWI, T2-DWI and complex mp models based on transfer learning (AUCs range from 0.76 to 0.81, 0.80 to 0.88, 0.85 to 0.92, and 0.94 to 0.96, respectively) outperformed the models based on radiomics (AUCs of 0.73, 0.76, 0.79, and 0.92, respectively). Moreover, the complex mp model showed the best prediction performance, with the Resnet50-complex mp model achieving the highest AUC (0.96) and accuracy (0.87).
Conclusions
Transfer learning is feasible and superior to radiomics in the differential diagnosis of uterine sarcomas and ALMs in our dataset. ML models based on deep learning features of nonenhanced mp-MRI and clinical parameters can achieve good diagnostic efficacy.
Key Points
• The ML model combining nonenhanced mp-MRI features and clinical parameters can distinguish uterine sarcomas from ALMs.
• Transfer learning can be applied to differentiate uterine sarcomas from ALMs and outperform radiomics.
• The most accurate prediction model was Resnet50-based transfer learning, built with the deep-learning features of mp-MRI and clinical parameters.




Similar content being viewed by others
Abbreviations
- ADC:
-
Apparent diffusion coefficient
- ALMs:
-
Atypical leiomyomas
- AUC:
-
Area under the curve
- CNN:
-
Convolutional neural network
- DWI:
-
Diffusion weighted imaging
- DCE-MRI:
-
Dynamic contrast-enhanced magnetic resonance imaging
- Grad-CAM:
-
Gradient-weighted class activation mapping
- LASSO:
-
Least absolute shrinkage selector operator
- ML:
-
Machine learning
- Mp-MRI:
-
Multiparametric magnetic resonance imaging
- RF:
-
Random forest
- ROC:
-
Receiver operating characteristic curve
- ROI:
-
Region of interest
- T2WI:
-
T2-weighted imaging
3.References
Bi Q, Xiao Z, Lv F, Liu Y, Zou C, Shen Y (2018) Utility of clinical parameters and multiparametric MRI as predictive factors for differentiating uterine sarcoma from atypical leiomyoma. Acad Radiol 25:993–1002
Tamai K, Koyama T, Saga T et al (2008) The utility of diffusion-weighted MR imaging for differentiating uterine sarcomas from benign leiomyomas. Eur Radiol 18:723–730
Mendez RJ (2020) MRI to differentiate atypical leiomyoma from uterine sarcoma. Radiology 297:372–373
Aleksandrovych V, Bereza T, Sajewicz M, Walocha J, Gil K (2015) Uterine fibroid: common features of widespread tumor. Folia Med Cracov 55
Yang S, Kong F, Hou R et al (2017) Ultrasound guided high-intensity focused ultrasound combined with gonadotropin releasing hormone analogue (GnRHa) ablating uterine leiomyoma with homogeneous hyperintensity on T 2 weighted MR imaging. Br J Radiol 90:20160760
Brito Pires NM, Godoi ET, Oliveira DC et al (2017) Impact of pelvic magnetic resonance imaging findings in the indication of uterine artery embolization in the treatment of myoma. Ginekol Pol 88:129–133
Owen C, Armstrong AY (2015) Clinical management of leiomyoma. Obstet Gynecol Clin North Am 42:67–85
Mbatani N, Olawaiye AB, Prat J (2018) Uterine sarcomas. Int J Gynaecol Obstet 143(Suppl 2):51–58
Namimoto T, Yamashita Y, Awai K et al (2009) Combined use of T2-weighted and diffusion-weighted 3-T MR imaging for differentiating uterine sarcomas from benign leiomyomas. Eur Radiol 19:2756–2764
Thomassin-Naggara I, Dechoux S, Bonneau C et al (2013) How to differentiate benign from malignant myometrial tumours using MR imaging. Eur Radiol 23:2306–2314
Abdel Wahab C, Jannot AS, Bonaffini PA et al (2020) Diagnostic algorithm to differentiate benign atypical leiomyomas from malignant uterine sarcomas with diffusion-weighted MRI. Radiology 297:361–371
Causa Andrieu P, Woo S, Kim TH, Kertowidjojo E, Hodgson A, Sun S (2021) New imaging modalities to distinguish rare uterine mesenchymal cancers from benign uterine lesions. Curr Opin Oncol 33:464–475
Malek M, Tabibian E, Dehgolan MR et al (2020) A diagnostic algorithm using multi-parametric MRI to differentiate benign from malignant myometrial tumors: machine-learning method. Sci Rep 10:1–12
Nakagawa M, Nakaura T, Namimoto T et al (2019) A multiparametric MRI-based machine learning to distinguish between uterine sarcoma and benign leiomyoma: comparison with (18)F-FDG PET/CT. Clin Radiol 74:167 e161-167 e167
Xie H, Hu J, Zhang X, Ma S, Liu Y, Wang X (2019) Preliminary utilization of radiomics in differentiating uterine sarcoma from atypical leiomyoma: comparison on diagnostic efficacy of MRI features and radiomic features. Eur J Radiol 115:39–45
Xie H, Zhang X, Ma S, Liu Y, Wang X (2019) Preoperative differentiation of uterine sarcoma from leiomyoma: comparison of three models based on different segmentation volumes using radiomics. Mol Imaging Biol 21:1157–1164
Nakagawa M, Nakaura T, Namimoto T et al (2019) Machine learning to differentiate T2-weighted hyperintense uterine leiomyomas from uterine sarcomas by utilizing multiparametric magnetic resonance quantitative imaging features. Acad Radiol 26:1390–1399
Liao D, Xiao Z, Lv F, Chen J, Qiu L (2020) Non-contrast enhanced MRI for assessment of uterine fibroids' early response to ultrasound-guided high-intensity focused ultrasound thermal ablation. Eur J Radiol 122:108670
Chan HP, Samala RK, Hadjiiski LM, Zhou C (2020) Deep learning in medical image analysis. Adv Exp Med Biol 1213:3–21
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444
Pan SJ, Yang Q (2010) A survey on transfer learning. IEEE Transactions on Knowledge and Data Engineering 22:1345–1359
Sharif Razavian A, Azizpour H, Sullivan J, Carlsson S (2014) CNN features off-the-shelf: an astounding baseline for recognition Proceedings of the IEEE conference on computer vision and pattern recognition workshops, pp 806-813
Liu W, Cheng Y, Liu Z et al (2021) Preoperative prediction of Ki-67 status in breast cancer with multiparametric MRI using transfer learning. Acad Radiol 28:e44–e53
Deepak S, Ameer PM (2019) Brain tumor classification using deep CNN features via transfer learning. Comput Biol Med 111:103345
Tian S, Niu M, Xie L, Song Q, Liu A (2021) Diffusion-tensor imaging for differentiating uterine sarcoma from degenerative uterine fibroids. Clin Radiol 76:313 e327-313 e332
van Griethuysen JJM, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77:e104–e107
Balagurunathan Y, Kumar V, Gu Y et al (2014) Test-retest reproducibility analysis of lung CT image features. J Digit Imaging 27:805–823
Hu Y, Xie C, Yang H et al (2021) Computed tomography-based deep-learning prediction of neoadjuvant chemoradiotherapy treatment response in esophageal squamous cell carcinoma. Radiother Oncol 154:6–13
Muthukrishnan R, Rohini R (2016) LASSO: A feature selection technique in predictive modeling for machine learning2016 IEEE international conference on advances in computer applications (ICACA). IEEE, pp 18-20
Gerges L, Popiolek D, Rosenkrantz AB (2018) Explorative investigation of whole-lesion histogram MRI metrics for differentiating uterine leiomyomas and leiomyosarcomas. AJR Am J Roentgenol 210:1172–1177
Truhn D, Schrading S, Haarburger C, Schneider H, Merhof D, Kuhl C (2019) Radiomic versus convolutional neural networks analysis for classification of contrast-enhancing lesions at multiparametric breast MRI. Radiology 290:290–297
Liu S, Zheng H, Feng Y, Li W (2017) Prostate cancer diagnosis using deep learning with 3D multiparametric MRIMedical imaging 2017: computer-aided diagnosis. International Society for Optics and Photonics, p 1013428
Lundervold AS, Lundervold A (2019) An overview of deep learning in medical imaging focusing on MRI. Z Med Phys 29:102–127
Takahashi M, Kozawa E, Tanisaka M, Hasegawa K, Yasuda M, Sakai F (2016) Utility of histogram analysis of apparent diffusion coefficient maps obtained using 3.0T MRI for distinguishing uterine carcinosarcoma from endometrial carcinoma. J Magn Reson Imaging 43:1301–1307
Testa AC, Di Legge A, Bonatti M, Manfredi R, Scambia G (2016) Imaging techniques for evaluation of uterine myomas. Best Pract Res Clin Obstet Gynaecol 34:37–53
Ghassemi M, Oakden-Rayner L, Beam AL (2021) The false hope of current approaches to explainable artificial intelligence in health care. Lancet Digit Health 3:e745–e750
Barral M, Place V, Dautry R et al (2017) Magnetic resonance imaging features of uterine sarcoma and mimickers. Abdom Radiol (NY) 42:1762–1772
Wang T, Gong J, Li Q et al (2021) A combined radiomics and clinical variables model for prediction of malignancy in T2 hyperintense uterine mesenchymal tumors on MRI. Eur Radiol 31:6125–6135
Xie H, Hu J, Zhang X, Ma S, Liu Y, Wang X (2019) Preliminary utilization of radiomics in differentiating uterine sarcoma from atypical leiomyoma: comparison on diagnostic efficacy of MRI features and radiomic features. Eur J Radiol 115:39–45
Chen I, Firth B, Hopkins L, Bougie O, Xie RH, Singh S (2018) Clinical characteristics differentiating uterine sarcoma and fibroids. JSLS 22
Acknowledgements
The author would like to thank Yang Liu for his contribution to the collection of patients. Thanks to Yan Hu, Guanghui Li, and Jian Zhang who participated in the discussion of this research together. Thanks to the public code provided by Yihuai Hu et al.
Funding
This study has received funding from Chongqing Postgraduate Research and Innovation Project (NO. CYB20165).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Fajin Lv.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• Retrospective
• diagnostic or prognostic study
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 250 kb)
Rights and permissions
About this article
Cite this article
Dai, M., Liu, Y., Hu, Y. et al. Combining multiparametric MRI features-based transfer learning and clinical parameters: application of machine learning for the differentiation of uterine sarcomas from atypical leiomyomas. Eur Radiol 32, 7988–7997 (2022). https://doi.org/10.1007/s00330-022-08783-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-022-08783-7