Abstract
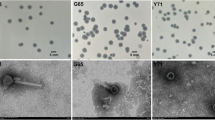
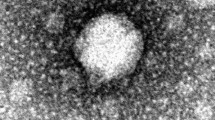
Aeromonas punctata is the causative agent of septicemia, diarrhea, wound infections, meningitis, peritonitis, and infections of the joints, bones and eyes. Bacteriophages are often considered alternative agents for controlling bacterial infection and contamination. In this study, we described the isolation and preliminary characterization of bacteriophage IHQ1 (family Myoviridae) active against the Gram-negative bacterial strain A. punctata. This virulent bacteriophage was isolated from stream water sample. Genome analysis indicated that phage IHQ1 was a double-stranded DNA virus with an approximate genome size of 25–28 kb. The initial characterization of this newly isolated phage showed that it has a narrow host range and infects only A. punctata as it failed to infect seven other clinically isolated pathogenic strains, i.e., methicillin-resistant Staphylococcus aureus 6403, MRSA 17644, Acinetobacter 33408, Acinetobacter 1172, Pseudomonas aeruginosa 22250, P. aeruginosa 11219, and Escherichia coli. Proteomic pattern of phage IHQ1, generated by SDS-PAGE using purified phage particles, showed three major and three minor protein bands with molecular weights ranging from 25 to 70 kDa. The adsorption rate of phage IHQ1 to the host bacterium was also determined, which was significantly enhanced by the addition of 10 mM CaCl2. From the single-step growth experiment, it was inferred that the latent time period of phage IHQ1 was 24 min and a burst size of 626 phages per cell. Moreover, the pH and thermal stability of phage IHQ1 were also investigated. The maximum stability of the phage was observed at optimal pH 7.0, and it was totally unstable at extreme acidic pH 3; however, it was comparatively stable at alkaline pH 11.0. At 37°C the phage showed maximum number of plaques, and the viability was almost 100%. The existence of Aeromonas bacteriophage is very promising for the eradication of this opportunistic pathogen and also for future applications such as the design of new detection and phage typing (diagnosis) methods. The specificity of the bacteriophage for A. punctata makes it an attractive candidate for phage therapy of A. punctata infections.










Similar content being viewed by others
References
Abedon S, Culler R (2007) Optimizing bacteriophage plaque fecundity. J Theor Biol 249:582–592
Abrams E, Zierdt CH, Brown JA (1971) Observations on Aeromonas hydrophila septicemia in a patient with leukemia. J Clin Pathol 24:491–492
Adams MH (1959) Bacteriophages. Interscience Publishers, Inc. New York
Ashelford KE, Norris SJ, Fry JC, Bailey MJ, Day MJ (2000) Seasonal population dynamics and interactions of competing bacteriophages and their host in the rhizosphere. Appl Environ Microbiol 66:4193–4199
Bohannan B, Lenski R (2000) Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol Lett 3:362–377
Bren L (2007) Bacteria-eating virus approved as food additive. FDA Consum 41:20–21
Bulger RJ, Sherris JC (1966) The clinical significance of Aeromonas hydrophila. Report of two cases. Arch Intern Med 118:562–564
Capra ML, Quiberoni A, Reinheimer J (2006) Phages of Lactobacillus casei/paracasei: response to environmental factors and interaction with collection and commercial strains. J Appl Microbiol 100:334–342
Capra ML, Quiberoni A, Reinheimer JA (2004) Thermal and chemical resistance of Lactobacillus casei and Lactobacillus paracasei bacteriophages. Lett Appl Microbiol 38:499–504
Carey-Smith GV, Billington C, Cornelius AJ, Hudson JA, Heinemann JA (2006) Isolation and characterization of bacteriophages infecting Salmonella spp. FEMS Microbiol Lett 258:182–186
Carlton RM, Noordman WH, Biswas B, De Meester ED, Loessner MJ (2005) Bacteriophage P100 for control of Listeria monocytogenes in foods: genome sequence, bioinformatics analyses, oral toxicity study, and application. Regul Toxicol Pharmacol 43:301–312
Casjens SR (2008) Diversity among the tailed-bacteriophages that infect the Enterobacteriaceae. Res Microbiol 159:340–348
Chow JJ, Batt CA, Sinskey AJ (1988) Characterization of Lactobacillus bulgaricus bacteriophage ch2. Appl Environ Microbiol 54:1138–1142
Chow MS, Rouf MA (1983) Isolation and partial characterization of two Aeromonas hydrophila bacteriophages. Appl Environ Microbiol 45:1670–1676
Clark JR, March JB (2006) Bacteriophages and biotechnology: vaccines, gene therapy and antibacterials. Trends Biotechnol 24:212–218
Davis WA, Kane JG, Garagusi VF (1978) Human Aeromonas infections: a review of the literature and a case report of endocarditis. Medicine (Baltimore) 57:267–277
DeFronzo RA, Murray GF, Maddrey WC (1973) Aeromonas septicemia from hepatobiliary disease. Am J Dig Dis 18:323–331
Eddy BP (1960) Cephalotrichous, fermentative Gram-negative bacteria: the genus Aeromonas. J Appl Bacteriol 23:216–249
Garrity G (2005) Bergey’s manual of systematic bacteriology. In: Garrity GM (ed) Bergey’s manual of systematic bacteriology, vol 2, 2nd edn. Williams and Wilkins, Philadelphia, pp 124–133
Gilbert P, Das J, Foley I (1997) Biofilm susceptibility to antimicrobials. Adv Dent Res 11:160–167
Hermoso JA, Garcia JL, Garcia P (2007) Taking aim on bacterial pathogens: from phage therapy to enzybiotics. Curr Opin Microbiol 10:461–472
Imbeault S, Parent S, Legace M, Uhland CF, Blais JF (2006) Using bacteriophages to prevent furunculosis caused by Aeromonas salmonicida in farmed brook trout. J Aqua Anim Health 18:203–214
Inal JM (2003) Phage therapy: a reappraisal of bacteriophages as antibiotics. Arch Immunol Ther Exp (Warsz) 51:237–244
Janda JM, Abbott SL (1998) Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations, and unanswered questions. Clin Infect Dis 27:332–344
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lenski R, Levin B (1985) Constraints on the coevolution of bacteria and virulent phage: a model, some experiments, and predictions for natural communities. Am Nat 125:585–602
Levin B, Bull J (2004) Population and evolutionary dynamics of phage therapy. Nat Rev Microbiol 2:166–173
Merril CR, Biswas B, Carlton R, Jensen NC, Creed GJ, Zullo S, Adhya S (1996) Long circulating bacteriophage as antibacterial agents. Proc Natl Acad Sci USA 93:3188–3192
Moldovan R, Chapman-McQuiston E, Wu X (2007) On kinetics of phage adsorption. Biophys J 93:303–315
Park SC, Shimarura I, Fukunaga M, Mori K, Nakai T (2000) Isolation of bacteriophages specific to a fish pathogen, Pseudomonas plecoglossicida, as a candidate for disease control. Appl Environ Microbiol 66:1416–1422
Popoff M (1971) Etude sur les Aeromonas salmonicida. II. Caracterisation des bacteriophages actifssur les “aeromonas salmonicida” etlysotypie. Ann Rech Vet 2:33–45
Puck T, Garen A, Cline J (1951) The mechanism of virus attachment to host cells. J Exp Med 93:65–88
Richards F (1969) The genetics of bacteria and their viruses. Yale J Biol Med 42:120–121
Shivu MM, Rajeeva BC, Girisha SK, Karunasagar I, Krohne G, Karunasagar I (2007) Molecular characterization of Vibrio harveyi bacteriophages isolated from aquaculture environments along the coast of India. Environ Microbiol 9:322–331
Stenholm AR, Dalsgaard I, Middelboe M (2008) Isolation and characterization of bacteriophages infecting the fish pathogen Flavobacterium psychrophilum. Appl Environ Microbiol 74:4070–4078
Su MT, Venkatesh TV, Bodmer R (1998) Large- and small-scale preparation of bacteriophage lysate and DNA. Biotechniques 25:44–46
Suárez V, Moineau S, Reinheimer J, Quiberoni A (2008) Argentinean Lactococcus lactis bacteriophages: genetic characterization and adsorption studies. J Appl Microbiol 104:371–379
Sulakvelidze A, Barrow P (2005) Phage therapy in animals and agribusiness. In: Kutter E, Sulakvelidze A (eds) Bacteriophages: biology and applications. CRC, Boca Raton, pp 335–380
Szczepanowski R, Bekel T, Goesmann A, Krause L, Krömeke H, Kaiser O, Eichler W, Pühler A, Schlüter A (2008) Insight into the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to antimicrobial drugs analysed by the 454-pyrosequencing technology. J Biotechnol 136:54–64
Wang I (2006) Lysis timing and bacteriophage fitness. Genetics 172:17–26
Wang I, Dykhuizen D, Slobodkin L (1996) The evolution of phage lysis timing. Evol Ecol 10:545–558
Watnick P, Kolter R (2000) Biofilm, city of microbes. J Bacteriol 182:2675–2679
Zahoor A, Rehman A (2009) Isolation of Cr(VI) reducing bacteria from industrial effluents and their potential use in bioremediation of chromium containing wastewater. J Environ Sci 21:814–820
Zimmer M, Scherer S, Loessner MJ (2002) Genomic analysis of Clostridium perfringens bacteriophage φ3626, which integrates into guaA and possibly affects sporulation. J Bacteriol 184:4359–4368
Acknowledgments
We would like to acknowledge Dr. Sohail Hamid and Mr. Javed Iqbal from the National Institute of Biotechnology and Genetic Engineering (NIBGE), Faisalabad, for transmission electron microscopy. We are thankful to Mr. Muhammad Shafique, Microbiology lab, Pakistan Institute of Medical Sciences (PIMS), Pakistan, for providing bacterial strains to carry out phage host range experiments. We are thankful to the Higher Education Commission (HEC) and the Ministry of Science and Technology (MoST), Pakistan, for providing funding to Dr. Ishtiaq Qadri.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Haq, I.U., Chaudhry, W.N., Andleeb, S. et al. Isolation and Partial Characterization of a Virulent Bacteriophage IHQ1 Specific for Aeromonas punctata from Stream Water. Microb Ecol 63, 954–963 (2012). https://doi.org/10.1007/s00248-011-9944-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-011-9944-2