Abstract
The main objective of this survey is to study the published articles to determine the most favorite data mining methods and gap of knowledge. Since the threat of pandemics has raised concerns for public health, data mining techniques were applied by researchers to reveal the hidden knowledge. Web of Science, Scopus, and PubMed databases were selected for systematic searches. Then, all of the retrieved articles were screened in the stepwise process according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses checklist to select appropriate articles. All of the results were analyzed and summarized based on some classifications. Out of 335 citations were retrieved, 50 articles were determined as eligible articles through a scoping review. The review results showed that the most favorite DM belonged to Natural language processing (22%) and the most commonly proposed approach was revealing disease characteristics (22%). Regarding diseases, the most addressed disease was COVID-19. The studies show a predominance of applying supervised learning techniques (90%). Concerning healthcare scopes, we found that infectious disease (36%) to be the most frequent, closely followed by epidemiology discipline. The most common software used in the studies was SPSS (22%) and R (20%). The results revealed that some valuable researches conducted by employing the capabilities of knowledge discovery methods to understand the unknown dimensions of diseases in pandemics. But most researches will need in terms of treatment and disease control.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Throughout history, the threat of pandemics has raised concerns for the healthcare community. The potential threat of spreading major infected diseases around the world before anyone aware of it is a controversial issue. The apparent prevalence of Severe Acute Respiratory Syndrome (SARS) and various types of influenza in the past have indicated the extent to which a pandemic disease can affect the health systems of countries [1, 2]. Coronavirus disease (COVID-19) is the last series of pandemic diseases that affect the world powerfully. COVID-19 or novel Coronavirus (2019-nCoV) is an infectious disease caused by coronavirus 2 (SARS-CoV-2) that began on December 8, 2019, from Wuhan, China [3, 4]. Since a novel coronavirus (nCoV) is a new strain of the coronavirus family that has not been seen before, the world faces serious challenges to control this outbreak [5, 6]. During the fierce outbreaks, not only clinical specialists have been trying to invent novel treatments and vaccines, but also scientists in the field of data science and technology are trying to discover the infectious and help control it by applying information-based methods [7, 8].
Nowadays, an extensive amount of health data is collected through patient care from different numerous sources due to the digital health revolution [9, 10]. Hence, the modern world of medicine is rich in information but it is poor in knowledge [11, 12]. Therefore, striving to this new pandemic and possible future pandemics has become one of the notable concerns of scientists.
In the last decades, some valuable studies have been published regarding pandemics and data mining (DM) techniques[13]. Such studies were conducted with the aim of better understanding, controlling, and manage pandemics using various data mining methods. Due to the importance to fight the COVID-19 pandemic, conducting a survey on the most popular and efficient data mining methods could have a significant impact on selecting the most effective techniques in pandemic studies. Thus, it can help us to reveal the unknown character of the new pandemic and the next possible pandemics. As follows, the core objective of this review is sought to collecting, summarizing, and analyzing the existing articles to aid track and analysis of such studies that have been published in terms of pandemics and data mining methods. The specific research questions (RQ) of this review are: (RQ1) To determine how many studies published over the past years and previous months regarding last pandemics and COVID-19 outbreak, (RQ2) Representing an overview of published studies and their characteristics, (RQ3) Investigating the published studies regarding data mining techniques, (RQ4) Identifying the source of data, (RQ5) Determining the most favorite DM techniques in terms of their frequency and clinical domains, (RQ6) Identifying the main approaches of published studies.
2 Method
The present study was completed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) checklist to ensure the inclusion of relevant studies [14]. Next, the synthesis of eligible articles based on the main characteristics was conducted to classify the main characteristics of studies.
2.1 Literature search
A systematic search of the scientific database, Web of Science, Scopus, and PubMed databases from 2010 up to 16 Oct 2020 was completed using “data mining”, “prediction model”, “data mining techniques”, “data mining methods”, “pandemics”, “pandemic”, “COVID-19”, “SARS-CoV-2”, and “coronavirus disease” as keywords. Boolean search strategies were designed based on these keywords in each database.
2.2 Inclusion and exclusion criteria for study selection
Articles were included if they met the following criteria:
1) The focus of this study is on pandemic diseases such as COVID-19,
2) Only the articles about using data mining techniques or knowledge discovery methods were included. Due to the variety of methods in this field, these types of methods are selected based on the study was conducted by Patel and Patel [15].
3) Studies were limited to those published in the English language.
Articles were excluded if they met the following criteria: 1) The title, abstract, or full text of the article did not relate to any pandemics or COVID-19 disease, 2) Book chapters, letters to editors, short briefs, reports, commentaries, technical reports, review or meta-analysis were excluded, 3) Non-English papers, 3) Image processing methods were not considered. 4) The full text was not available. To reduce the bias of unavailable full-text, the full texts of non-open access articles were obtained by contacting to authors. Therefore, all of the full-text of articles were retrieved by researchers.
2.3 Data extraction phase
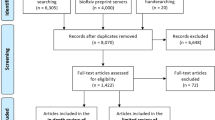
In scientific databases searching (Web of Science, Scopus, and PubMed), 311 articles were retrieved through the web interface of scientific websites. Some inclusion and exclusion criteria were defined for screening papers. In the first phase, all titles and abstracts of retrieved articles were examined to select eligible studies. All of the titles and abstracts were screened by three reviewers (MT, SS, and SR) to find relevant articles. Another reviewer (MG) reviewed a sample of studies randomly. The quality analysis of the individual papers was assessed by the Joanna Briggs Institute (JBI) checklist which provides robust checklists for the appraisal and assessment of most types of studies [16, 17]. Since all types of studies were included in our review, we applied this checklist. Decisions on study eligibility and quality were made by two reviewers; any disagreements were resolved by discussion. The flow of screening articles based on the [17] PRISMA method illustrates in Fig. 1.
Phase three involves full-text screening. In this phase, the full texts of relevant studies were screened thoroughly by four reviewers (MT, SS, SR, and MG). Through a full-text review, the final decision was made by RS if there was a disagreement between the authors in the selection of eligible studies.
Finally, 50 studies remained as eligible articles. Some classifications were assumed to classify and analyze the included studies. The extraction forms were designed by researchers to manage the reviewed articles. This classification comprises general information and specific information. General information includes author names, publication date, and publisher. Specific information includes the main objective, DM techniques, application of DM method, health discipline, main outcomes, evaluation results, data sources, sample sizes, applied software, and country. Included articles were analyzed to extract their characteristics based on the predefined classification. All of the extracted information was re-examined by all authors to reach an agreement. The next reviewer (RS) evaluated and validated the results. EndNote X9 is used for resource management, and all qualitative analysis was performed in SPSS v20.
3 Results
Earlier searches in scientific databases yielded 311 citations. First, 13 articles were excluded in the duplicate removal phase. Next, 82 articles were omitted due to their irrelevancy in the full-text screening stage. All included articles could be included in our review according to the JBI checklist. In the last screening phase. Finally, 50 articles were identified as eligible studies.
3.1 Study characteristics
All eligible papers that met our inclusion criteria included 47 journal papers and three conference papers. The distribution of studies by year is described in Table 1. As it is apparent, the majority of studies were published in 2020. Thus, the frequency of publication of these articles by month in 2020 was also examined. The trend of published articles regarding the month in 2020 is shown in Fig. 2. The “International Journal of Environmental Research and Public Health” has the first rank with six articles among the journals.
A summary of the included articles based on predefined categories is described in Table 4 in Appendix. To visualize the frequency of words that appeared more frequently in reviewed articles, all articles summarized in the word cloud in Fig. 3.
3.2 Sample size and data sources of articles
Out of 50 studies, only 35 citations reported their sample size. Due to the variety of samples, the range of sample size was very wide. In other words, the samples considered were very different due to the variety of applied methods. The sample size ranged from 53 cases to 1,413,297 posts. In total, 35 different data sources were cited for eligible articles. social media platforms (n = 10), Hospital information sources (n = 7), and World Health Organization (n = 4) data sources were the three most common sources of information.
3.3 The distribution of articles based on the countries
In terms of the country, articles have been published in 14 different countries. The article also uses global data on the disease pandemic. The distribution of articles by country is shown in Fig. 4 on the worldwide map. As it turns out, China has the highest frequency among other countries.
3.4 The distribution of literature by main approaches
All of the articles in this study took a specific approach to fight the pandemic diseases and provide a better understanding by applying DM techniques. Based on the survey, we classified all of the articles by their main approaches in 11 categories that are shown in Table 2. One of the main objectives of eligible articles is Infoveillance. The term infoveillance has come to be used to refer to a type of syndromic surveillance that uses information and online tools in public health domains. Regarding infoveillance, regression was applied to provide new insight into the origins of the outbreak based on the analysis of social media information [18].
As can be seen from Table 2, the majority of studies (22%) devoted to the disease characteristic. In the case of diseases, studies show that the most common use of data mining techniques to fight pandemics was related to the new pandemic COVID-19 (n = 44). Other diseases such as H1N1 Influenza (n = 2), Other types of Influenza pandemics (n = 2), and SARS (n = 2) were also considered.
3.5 The distribution of data mining techniques in reviewed articles
Since the main objective of this study was to determine to what extent data mining techniques are employed to fight pandemics, the frequency of applied methods was investigated in this section according to a study conducted by Patel and Patel [15]. Table 3 showed an overview of the distribution of applied data mining methods in reviewed articles. The analysis showed that all of the applied methods were classified into 14 main categories. It is apparent that the most favorite method was employed in reviewed articles belonged to Natural language processing (NLP) techniques (22%). While logistic regression analysis with 20% of studies was in the second rank to determine the association of the independent variables with one dichotomous dependent variable[68]. It should be noted here that most studies have used more than one data mining technique.
Additionally, the distribution of employed DM techniques regarding main approaches is illustrated in Fig. 5. The distribution and frequency of employed DM techniques based on main approaches can provide an appropriate insight for researchers regarding pandemics. The numbers in this figure indicate the number of studies per axis.
All of the DM techniques are categorized into supervised and unsupervised techniques. In a supervised learning method, the algorithm learns on a labeled dataset to provide an answer. While unsupervised learning techniques in which patterns are extracted from the unlabeled input data [69]. Thus, all of the applied methods in the reviewed articles were divided into three categories: supervised techniques (90%), unsupervised techniques (4%), and a combination of supervised and unsupervised techniques (6%).
3.6 The distribution of reviewed articles based on applied software
Special tools and a suitable platform are needed to perform data mining methods. In this section, we have examined the frequency of various tools used in these studies. SPSS software has the highest percentage (22%) among other tools, next R software has the second rank with 10 papers (20%), followed by Python software with nine studies (18%). MATLAB and RapidMiner software also accounted for one percent of the studies. Out of 50 studies, 13 studies (26%) did not specify the employed tools.
3.7 The characteristics of reviewed articles based on the main health domains
According to reviewed studies, we can classify all eligible articles in this review into eight categories based on their clinical discipline. The identified clinical and health disciplines with their distribution and their frequency are described in Fig. 6. From the chart, it is obvious that the greatest demand belonged to infectious disease with 18 papers (36%). Next, epidemiology is the second most discipline considered by included studies with 13 studies (26%). This analysis can be highly useful to determine literature gaps in terms of health domains.
4 Discussion
The main objective of this review was to summarize the studies carried out on the application of data-driven DM methods in pandemics. Therefore, 50 articles were selected and analyzed from 311 retrieved studies. The finding and results are discussed in this section. The data sources used in the included studies were very diverse. In terms of country, most studies were conducted in China. This can be explained by the fact that most pandemics began in this country.
Nowadays, social media has become a new source of data [70] and they can generate more information in a short period than other resources. Since accessibility to these kinds of data is easier than other sources of data, the foremost of studies were devoted to applying text mining techniques regarding Infoveillance. The qualitative analysis revealed that researchers preferred to use supervised techniques such as regression to produce predictive models for a better understanding of unknown pandemics. All of these methods have been pragmatically used in different fields of medicine efficiently [71]. Additionally, classification methods have been used more than predicted in studies. By selecting the best method for implementing accurate prediction models, researchers can discover certain biomarkers in unknown diseases which can allow them to forecast important outcomes [72, 73]. Therefore, developing prediction models not only can help physicians but also aid health policymakers and societies.
Since the majority of studies were conducted in China, these models may be faced with overfitting. However, none of the studies recommended applying developed models in real practice. However, most authors were optimistic about the development of predictive models. Shamsuddin's opinion regarding the development of forecasting models is in line with our study[74]. Wyntass et al. conducted a systematic review study regarding predictive models of COVID-19. They concluded that proposed models are poorly reported with a high risk of bias [75].
Results showed that controlling the transmission of infectious disease is the main concern in pandemic disease [76]. Usually, the nature of a new disease in a pandemic is unknown, and identifying the characteristics of a new disease is one of the most important concerns for scientists. That it’s why the majority of studies are devoted to revealing disease characteristics. It can be explained by the fact that scientists should be paid more attention to diagnosis than other tasks in pandemic disease [77]. The next important issue in pandemic diseases is how the disease spreads. Hence, almost 10% of the studies have been dedicated to predicting the prevalence of the disease.
However, the sample size of datasets is very diverse due to a variety of applied methods. The results showed that most of the studies used various data sources with a limited number of data sets. Using large data sets can improve the strength of the results and improve the accuracy of the model's predictions [78], which in turn can help scientists better to fight this new disease. Accordingly, researchers are recommended to use large datasets for their studies even internationally, to achieve better diagnostic and therapeutic decisions.
In terms of diseases, most efforts were made under the heading of COVID-19. In the second place, the topics were related to influenza pandemics. This result is expected due to the high prevalence of these two diseases. Using and retrieving large amounts of data provided by electronic systems as a data source can improve access to data [79]. As a result, conducting data-driven studies has become easier in recent years than ever before. The fact that diseases related to other pandemics did not appear in this search may be due to the authors of these articles considered these diseases as epidemics.
In this study, we encountered some limitations. Nowadays, a vast majority of studies are published regarding COVID-19 daily. We investigated the literature up to 16 Oct 2020. Therefore, some studies might be neglected in the publication time of this article. Consequently, further research is needed to complete our results. Another limitation of the proposed research is that the electronic search process was performed in only three journal databases, and the rest of the databases were skipped while accessing the quality of journal articles which can be addressed in future research. The present study helps researchers to have a useful background for future work to understand the general context of data mining techniques in pandemics and their applications. Further studies could cover the study of data mining applications in a broader concept, or it can include the development of search strategies in larger databases. Analyzing and incorporating non-English written papers with automatic translator tools could be the subject of the next article. At least, it could be interesting to compare the number of non-English papers with English ones.
5 Conclusion
This review could help scientists to reach published researches regarding DM techniques and fierce pandemics easier. In this study, we surveyed the data mining techniques utilized in global pandemics, however, most of these techniques have been developed in the current context to prevent and predict the COVID-19 epidemic. According to our survey, we found out that the foremost objective of DM applications is related to disease characteristics. Also, it can help the policymakers and decision-makers in better decision-making regarding managing and preventing the major pandemics in the countries.
Data Availability
The study involves only a review of the literature without involving any data.
References
Jain V, Duse A, Bausch DG. Planning for large epidemics and pandemics: challenges from a policy perspective. Curr Opin Infect Dis. 2018;31(4):316–24.
Cook AH, Cohen DB. Pandemic Disease: A Past and Future Challenge to Governance in the United States. Rev Policy Res. 2008;25(5):449–71. https://doi.org/10.1111/j.1541-1338.2008.00346.x.
Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Di Napoli R. Features, evaluation, and treatment coronavirus (COVID-19). StatPearls [Internet]. StatPearls Publishing 2020.
Cucinotta D, Vanelli M. WHO declares COVID-19 a pandemic. Acta bio-medica: Atenei Parmensis. 2020; 91(1): 157–60.
Meo SA, Al-Khlaiwi T, Usmani AM, Meo AS, Klonoff DC, Hoang TD. Biological and Epidemiological Trends in the Prevalence and Mortality due to Outbreaks of Novel Coronavirus COVID-19. Journal of King Saud University - Science. 2020. https://doi.org/10.1016/j.jksus.2020.04.004.
Zhang S, Diao M, Yu W, Pei L, Lin Z, Chen D. Estimation of the reproductive number of novel coronavirus (COVID-19) and the probable outbreak size on the Diamond Princess cruise ship: A data-driven analysis. Int J Infect Dis. 2020; 93: 201–4. https://doi.org/10.1016/j.ijid.2020.02.033.
Atif I, Cawood FT, Mahboob MA. The Role of Digital Technologies that Could Be Applied for Prescreening in the Mining Industry During the COVID-19 Pandemic. Transactions of the Indian National Academy of Engineering. 2020:1–12. https://doi.org/10.1007/s41403-020-00164-0.
Gholamzadeh M, Abtahi H, Safdari R. Suggesting a framework for preparedness against the pandemic outbreak based on medical informatics solutions: a thematic analysis. The International Journal of health planning and management. 2021; n/a(n/a). https://doi.org/10.1002/hpm.3106.
Salzberger B, Glück T, Ehrenstein B. Successful containment of COVID-19: the WHO-Report on the COVID-19 outbreak in China. Springer; 2020.
Sun K, Chen J, Viboud C. Early epidemiological analysis of the coronavirus disease 2019 outbreak based on crowdsourced data: a population-level observational study. The Lancet Digital Health. 2020; 2(4): e201–8. https://doi.org/10.1016/S2589-7500(20)30026-1.
Heymann DL, Shindo N. COVID-19: what is next for public health? The Lancet. 2020; 395(10224): 542–5.
Keesara S, Jonas A, Schulman K. Covid-19 and Health Care’s Digital Revolution. N Engl J Med. 2020. https://doi.org/10.1056/NEJMp2005835.
Gulyaeva M, Huettmann F, Shestopalov A, Okamatsu M, Matsuno K, Chu D-H, et al. Data mining and model-predicting a global disease reservoir for low-pathogenic Avian Influenza (A) in the wider pacific rim using big data sets. Sci Rep. 2020; 10(1): 16817-. https://doi.org/10.1038/s41598-020-73664-2.
Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ (Clinical research ed). 2009; 339: b2535. https://doi.org/10.1136/bmj.b2535.
Patel S, Patel H. Survey of Data Mining Techniques used in Healthcare Domain. International Journal of Information Sciences and Techniques. 2016; 6(1/2): 53–60. https://doi.org/10.5121/ijist.2016.6206.
Institute JB. The Joanna Briggs Institute Critical Appraisal Tools. University of Adelaide, South Australia. 2017. https://jbi.global/critical-appraisal-tools. Accessed 5 Mar 2021.
Hannes K, Lockwood C, Pearson A. A comparative analysis of three online appraisal instruments’ ability to assess validity in qualitative research. Qual Health Res. 2010; 20(12): 1736–43. https://doi.org/10.1177/1049732310378656.
Li J, Xu Q, Cuomo R, Purushothaman V, Mackey T. Data Mining and Content Analysis of Chinese Social Media Platform Weibo During Early COVID-19 Outbreak: A Retrospective Observational Infoveillance Study. JMIR Public Health Surveill. 2020. https://doi.org/10.2196/18700.
Ahamad MM, Aktar S, Rashed-Al-Mahfuz M, Uddin S, Lio P, Xu HM, et al. A machine learning model to identify early stage symptoms of SARS-Cov-2 infected patients. Expert Systems with Applications. 2020; 160. https://doi.org/10.1016/j.eswa.2020.113661.
Huang C, Xu X, Cai Y, Ge Q, Zeng G, Li X, et al. Mining the characteristics of COVID-19 patients in China: Analysis of social media posts. J Med Internet Res. 2020; 22(5). https://doi.org/10.2196/19087.
Maram B, Padmapriya G, Satish AR. A framework for performance analysis on machine learning algorithms using covid-19 dataset. Adv Math: Sci J. 2020; 9(10): 8207–15. https://doi.org/10.37418/amsj.9.10.50.
Foieni F, Sala G, Mognarelli JG, Suigo G, Zampini D, Pistoia M, et al. Derivation and validation of the clinical prediction model for COVID-19. Intern Emerg Med. 2020. https://doi.org/10.1007/s11739-020-02480-3.
Ma XX, Li A, Jiao MF, Shi QM, An XC, Feng YH, et al. Characteristic of 523 COVID-19 in Henan Province and a Death Prediction Model. Frontiers in Public Health. 2020; 8. https://doi.org/10.3389/fpubh.2020.00475.
Luo Y, Mao LY, Yuan X, Xue Y, Lin Q, Tang GX, et al. Prediction Model Based on the Combination of Cytokines and Lymphocyte Subsets for Prognosis of SARS-CoV-2 Infection. J Clin Immunol. 2020; 40(7): 960–9. https://doi.org/10.1007/s10875-020-00821-7.
Ciucurel C, Iconaru EI. An Epidemiological Study on the Prevalence of the Clinical Features of SARS-CoV-2 Infection in Romanian People. Int J Environ Res Public Health. 2020; 17(14). https://doi.org/10.3390/ijerph17145082.
Roland LT, Gurrola JG, Loftus PA, Cheung SW, Chang JLL. Smell and taste symptom-based predictive model for COVID-19 diagnosis. International Forum of Allergy & Rhinology. 2020;10(7):832–8. https://doi.org/10.1002/alr.22602.
Liu Q, Song NC, Zheng ZK, Li JS, Li SK. Laboratory findings and a combined multifactorial approach to predict death in critically ill patients with COVID-19: a retrospective study. Epidemiology and Infection. 2020; 148. https://doi.org/10.1017/s0950268820001442.
Li D, Chaudhary H, Zhang Z. Modeling Spatiotemporal Pattern of Depressive Symptoms Caused by COVID-19 Using Social Media Data Mining. Int J Environ Res Public Health. 2020; 17(14). https://doi.org/10.3390/ijerph17144988.
Sarker A, Lakamana S, Hogg-Bremer W, Xie A, Al-Garadi MA, Yang YC. Self-reported COVID-19 symptoms on Twitter: an analysis and a research resource. J Am Med Inform Assoc. 2020; 27(8): 1310–5. https://doi.org/10.1093/jamia/ocaa116.
Abd-Alrazaq A, Alhuwail D, Househ M, Hamdi M, Shah Z. Top Concerns of Tweeters During the COVID-19 Pandemic: Infoveillance Study. J Med Internet Res. 2020;22(4):e19016. https://doi.org/10.2196/19016.
Zhang Y, Cheng J, Yang Y, Li H, Zheng X, Chen X, et al. Covid-19 public opinion and emotion monitoring system based on time series thermal new word mining. Comput Mater Continua. 2020; 64(3): 1415–34. https://doi.org/10.32604/cmc.2020.011316.
Han X, Wang J, Zhang M, Wang X. Using social media to mine and analyze public opinion related to COVID-19 in China. Int J Environ Res Public Health. 2020; 17(8). https://doi.org/10.3390/ijerph17082788.
Qin L, Sun Q, Wang Y, Wu KF, Chen M, Shia BC, et al. Prediction of number of cases of 2019 novel coronavirus (COVID-19) using social media search index. Int J Environ Res Public Health. 2020; 17(7). https://doi.org/10.3390/ijerph17072365.
Ayyoubzadeh SM, Ayyoubzadeh SM. Predicting COVID-19 Incidence Through Analysis of Google Trends Data in Iran: Data Mining and Deep Learning Pilot Study. JMIR Public Health Surveill. 2020; 6(2): e18828. https://doi.org/10.3855/jidc.12585/10.2196/18828.
Kostoff RN. Literature-related discovery: potential treatments and preventatives for SARS. Technol Forecast Soc Chang. 2011;78(7):1164–73.
Szomszor M, Kostkova P, St Louis C, editors. Twitter informatics: tracking and understanding public reaction during the 2009 swine flu pandemic. 2011 IEEE/WIC/ACM International Conferences on Web Intelligence and Intelligent Agent Technology; 2011: IEEE.
Wahbeh A, Nasralah T, Al-Ramahi M, El-Gayar O. Mining Physicians’ Opinions on Social Media to Obtain Insights Into COVID-19: Mixed Methods Analysis. JMIR Public Health Surveill. 2020; 6(2): e19276. https://doi.org/10.2196/19276.
Chintalapudi N, Battineni G, Amenta F. COVID-19 virus outbreak forecasting of registered and recovered cases after sixty day lockdown in Italy: A data driven model approach. Journal of Microbiology, Immunology and Infection. 2020; InPress. https://doi.org/10.1016/j.jmii.2020.04.004.
Khan MA, Abbas S, Khan KM, Al Ghamdi MA, Rehman A. Intelligent Forecasting Model of COVID-19 Novel Coronavirus Outbreak Empowered with Deep Extreme Learning Machine. Cmc-Computers Materials & Continua. 2020; 64(3): 1329–42. https://doi.org/10.32604/cmc.2020.011155.
Kargarfard F, Sami A, Hemmatzadeh F, Ebrahimie E. Identifying mutation positions in all segments of influenza genome enables better differentiation between pandemic and seasonal strains. Gene. 2019;697:78–85. https://doi.org/10.1016/j.gene.2019.01.014.
Kostkova P, Szomszor M, St. Louis C. Swineflu: The use of twitter as an early warning and risk communication tool in the 2009 swine flu pandemic. ACM Transactions on Management Information Systems (TMIS). 2014; 5(2): 1–25.
Dong YL, Zhou HF, Li MY, Zhang ZL, Guo WN, Yu T, et al. A novel simple scoring model for predicting severity of patients with SARS-CoV-2 infection. Transboundary and Emerging Dis. 2020. https://doi.org/10.1111/tbed.13651.
Verity R, Okell LC, Dorigatti I, Winskill P, Whittaker C, Imai N, et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis. The Lancet Infectious Dis. 2020; InPress. https://doi.org/10.1016/S1473-3099(20)30243-7.
Ghosal S, Sinha B, Majumder M, Misra A. Estimation of effects of nationwide lockdown for containing coronavirus infection on worsening of glycosylated haemoglobin and increase in diabetes-related complications: A simulation model using multivariate regression analysis. Diabetes Metab Syndr. 2020;14(4):319–23. https://doi.org/10.1016/j.dsx.2020.03.014.
Cheng FY, Joshi H, Tandon P, Freeman R, Reich DL, Mazumdar M, et al. Using Machine Learning to Predict ICU Transfer in Hospitalized COVID-19 Patients. Journal of Clinical Medicine. 2020; 9(6). https://doi.org/10.3390/jcm9061668.
Zhao ZR, Chen AN, Hou W, Graham JM, Li HF, Richman PS, et al. Prediction model and risk scores of ICU admission and mortality in COVID-19. Plos One. 2020; 15(7). https://doi.org/10.1371/journal.pone.0236618.
Alzahrani SI, Aljamaan IA, Al-Fakih EA. Forecasting the spread of the COVID-19 pandemic in Saudi Arabia using ARIMA prediction model under current public health interventions. J Infect Public Health. 2020; 13(7) :914–9. https://doi.org/10.1016/j.jiph.2020.06.001.
Anastassopoulou C, Russo L, Tsakris A, Siettos C. Data-based analysis, modelling and forecasting of the COVID-19 outbreak. PLoS ONE. 2020; 15(3). https://doi.org/10.1371/journal.pone.0230405.
Pinter G, Felde I, Mosavi A, Ghamisi P, Gloaguen R. COVID-19 Pandemic Prediction for Hungary; A Hybrid Machine Learning Approach. Mathematics. 2020; 8(6). https://doi.org/10.3390/math8060890.
Qiang XL, Xu P, Fang G, Liu WB, Kou Z. Using the spike protein feature to predict infection risk and monitor the evolutionary dynamic of coronavirus. Infect Dis Poverty. 2020; 9(1) :33. https://doi.org/10.1093/cid/ciaa3221186/s40249-020-00649-8.
Zhou YW, He YQ, Yang H, Yu H, Wang T, Chen Z, et al. Development and validation a nomogram for predicting the risk of severe COVID-19: A multi-center study in Sichuan, China. Plos One. 2020; 15(5). 10.1371/journal.pone.0233328.
Yan L, Zhang HT, Goncalves J, Xiao Y, Wang ML, Guo YQ et al. An interpretable mortality prediction model for COVID-19 patients. Nature Machine Intelligence. 2020; 2(5): 283-+. https://doi.org/10.1038/s42256-020-0180-7.
Jiang X, Coffee M, Bari A, Wang J, Jiang X, Huang J, et al. Towards an Artificial Intelligence Framework for Data-Driven Prediction of Coronavirus Clinical Severity. Computers, Materials \& Continua. 2020; 63(1). https://doi.org/10.32604/cmc.2020.010691.
Li S, Wang Y, Xue J, Zhao N, Zhu T. The impact of COVID-19 epidemic declaration on psychological consequences: a study on active Weibo users. Int J Environ Res Public Health. 2020; 17(6): 2032.
Martin-Rodriguez F, Sanz-Garcia A, Lopez-Izquierdo R, Benito JFD, Martin-Conty JL, Villamor MAC, et al. Predicting Health Care Workers’ Tolerance of Personal Protective Equipment: An Observational Simulation Study. Clin Simul Nurs. 2020; 47: 65–72. https://doi.org/10.1016/j.ecns.2020.07.005.
Ketu S, Mishra PK. Enhanced Gaussian process regression-based forecasting model for COVID-19 outbreak and significance of IoT for its detection. Appl Intell. 2020. https://doi.org/10.1007/s10489-020-01889-9.
Moftakhar L. The Exponentially Increasing Rate of Patients Infected with COVID-19 in Iran. Archives of Iranian medicine. 2020; 23(4): 235–8. https://doi.org/10.34172/aim.2020.0534172/aim.2020.03
Yongjian Z, Jingu X, Fengming H, Liqing C. Association between short-term exposure to air pollution and COVID-19 infection: Evidence from China. Sci Total Environ. 2020: 138704. https://doi.org/10.1016/j.scitotenv.2020.138704.
Sudirman ID, Nugraha DY. Naive Bayes classifier for predicting the factors that influence death due to covid-19 in China. J Theor Appl Inf Technol. 2020; 98(10): 1686–96.
Sudirman ID, Aryanto R, Mulyani. Optimizing decision tree criteria for predicting COVID-19 mortality in South Korea dataset. J Theor Appl Inf Technol. 2020; 98(15): 2889–900.
Fan Q, Zhu HL, Zhao JX, Zhuang LF, Zhang H, Xie HY. Risk factors for myocardial injury in patients with coronavirus disease et al 2019 in China Esc Heart Failure 2020 https://doi.org/10.1002/ehf2.13022
Ibrahim S, Kamaruddin SA, Sabri N, Samah KA, Noordin M, Shari A. The influences of global geographical climate towards COVID-19 spread and death. Int J Adv Trends Comput Sci Eng. 2020; 9(1.4 Special Issue): 612–7. https://doi.org/10.30534/ijatcse/2020/8591.42020.
Lei MT, Monjardino J, Mendes L, Goncalves D, Ferreira F. Statistical Forecast of Pollution Episodes in Macao during National Holiday and COVID-19. Int J Environ Res Public Health. 2020;17(14). https://doi.org/10.3390/ijerph17145124.
Ren X, Shao XX, Li XX, Jia XH, Song T, Zhou WY, et al. Identifying potential treatments of COVID-19 from Traditional Chinese Medicine (TCM) by using a data-driven approach. J Ethnopharmacol. 2020; 258. https://doi.org/10.1016/j.jep.2020.112932.
Neuraz A, Lerner I, Digan W, Paris N, Tsopra R, Rogier A, et al. Natural Language Processing for Rapid Response to Emergent Diseases: Case Study of Calcium Channel Blockers and Hypertension in the COVID-19 Pandemic. J Med Internet Res. 2020; 22(8): e20773. https://doi.org/10.2196/20773.
Masand VH, Rastija V, Patil MK, Gandhi A, Chapolikar A. Extending the identification of structural features responsible for anti-SARS-CoV activity of peptide-type compounds using QSAR modelling. SAR QSAR Environ Res. 2020; 31(9): 643–54. https://doi.org/10.1080/1062936x.2020.1784271.
Kargarfard F, Sami A, Ebrahimie E. Knowledge discovery and sequence-based prediction of pandemic influenza using an integrated classification and association rule mining (CBA) algorithm. J Biomed Inform. 2015; 57: 181–8. https://doi.org/10.1016/j.jbi.2015.07.018.
Han J, Pei J, Kamber M. Data Mining: Concepts and Techniques. Elsevier Science 2011.
Deo RC. Machine Learning in Medicine. Circulation. 2015; 132(20): 1920–30. https://doi.org/10.1161/CIRCULATIONAHA.115.001593.
Asur S, Huberman BA, editors. Predicting the future with social media. 2010 IEEE/WIC/ACM international conference on web intelligence and intelligent agent technology; 2010: IEEE.
Zhang Y, Guo SL, Han LN, Li TL. Application and Exploration of Big Data Mining in Clinical Medicine. Chin Med J. 2016;129(6):731–8. https://doi.org/10.4103/0366-6999.178019.
Alanazi HO, Abdullah AH, Qureshi KN, Ismail AS. Accurate and dynamic predictive model for better prediction in medicine and healthcare. Irish Journal of Medical Science (1971 -). 2018; 187(2):501–13. https://doi.org/10.1007/s11845-017-1655-3.
Nithya B, Ilango V, editors. Predictive analytics in health care using machine learning tools and techniques. 2017 International Conference on Intelligent Computing and Control Systems (ICICCS); 2017 15–16 June 2017.
Shamsoddin E. Can medical practitioners rely on prediction models for COVID-19? A systematic review Evidence-based dentistry. 2020;21(3):84–6.
Wynants L, Van Calster B, Collins GS, Riley RD, Heinze G, Schuit E, et al. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ (Clinical research ed). 2020; 369: m1328-m. https://doi.org/10.1136/bmj.m1328.
Adhikari SP, Meng S, Wu Y-J, Mao Y-P, Ye R-X, Wang Q-Z, et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect Dis Poverty. 2020;9(1):29. https://doi.org/10.1186/s40249-020-00646-x.
Kelly-Cirino CD, Nkengasong J, Kettler H, Tongio I, Gay-Andrieu F, Escadafal C, et al. Importance of diagnostics in epidemic and pandemic preparedness. BMJ global health. 2019; 4(Suppl 2): e001179-e. https://doi.org/10.1136/bmjgh-2018-001179.
Fortuny EJd, Martens D, Provost F. Predictive Modeling With Big Data: Is Bigger Really Better? Big Data. 2013; 1(4): 215–26. https://doi.org/10.1089/big.2013.0037.
Sun W, Cai Z, Li Y, Liu F, Fang S, Wang G. Data Processing and Text Mining Technologies on Electronic Medical Records: A Review. Journal of Healthcare Engineering. 2018: 4302425. https://doi.org/10.1155/2018/4302425.
Mudunuri SB, Nagarajaram H, Mishra P, editors. Distributional analysis and motif frequencies of compound microsatellite repeats in viral genomes. 2012 International Conference on Computing, Electronics and Electrical Technologies (ICCEET); 2012: IEEE.
Funding
The author(s) received no financial support for the research, or publication of this article.
Author information
Authors and Affiliations
Contributions
M.Gholamzadeh, S. Saeedi, S. Rezayi, and M.Tanhapour designed the systematic review, search strategy, and conducted database searches. M.Gholamzadeh, S.Saeedi, S.Rezayi, and M.Tanhapour conducted article screenings under R.Safdari supervision. M.Gholamzadeh conducted the analysis and interpretation under R.Safdari's supervision. M.Gholamzadeh S. Saeedi, S. Rezayi, and M.Tanhapour drafted the manuscript. All authors reviewed the content.
Corresponding author
Ethics declarations
Ethics statement
The study involves only a review of literature without involving humans and/or animals. The authors have no ethical conflicts to disclose.
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A
Appendix A
Rights and permissions
About this article
Cite this article
Safdari, R., Rezayi, S., Saeedi, S. et al. Using data mining techniques to fight and control epidemics: A scoping review. Health Technol. 11, 759–771 (2021). https://doi.org/10.1007/s12553-021-00553-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12553-021-00553-7