Abstract
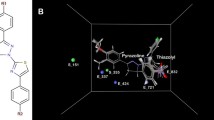
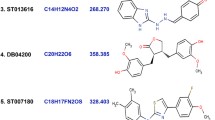
Human lemur tyrosine kinase-3 (LMTK3) is an oncogenic kinase known to regulate ER-α through phosphorylation and is considered to be a novel therapeutic target for breast cancer. In this work, we have studied the ATP-binding mechanism with LMTK3 domain and also carried out virtual screening on LMTK3 to identify lead compounds using Dock blaster server. The top scored compounds obtained from Dock blaster were then narrowed down further to six lead compounds (ZINC37996511, ZINC83363046, ZINC3745998, ZINC50456700, ZINC83351792 and ZINC83364581) based on high-binding affinity and non-bonding interactions with LMTK3 using Autodock 4.2 program. We found in comparison to ATP, the lead compounds bind relatively stronger to LMTK3. The relative binding free energy results from MM-PBSA/GBSA method further indicate the strong binding affinity of lead compounds over ATP to LMTK3 in the dynamic system. Further, potential of mean force (PMF) study for ATP and lead compounds with LMTK3 have been performed to explore the unbinding processes and the free energy barrier. From the PMF results, we observed that the lead compounds have higher dissociation energy barriers than the ATP. Our findings suggest that these lead compounds may compete with ATP, and could act as probable potential inhibitors for LMTK3.









Similar content being viewed by others
References
Albrand G, Terret C (2008) Early breast cancer in the elderly: assessment and management considerations. Drug Aging 25:35–45. https://doi.org/10.2165/00002512-200825010-00004
Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstrin NS, Hayes M, Hicks DG, Lester S, Love R, Mangu PB, McShane L (2010) American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of and progesterone receptors in breast cancer. J Clin Oncol 28:2784–2795. https://doi.org/10.1200/jco.2009.25.6529
Colditz GA (1998) Relationship between estrogen levels, use of hormone replacement therapy, and breast cancer. J Natl Cancer Inst 90:814–823. https://doi.org/10.1093/jnci/90.11.814
Hankinson SE, Colditz GA, Willett WC (2004) Towards an integrated model for breast cancer etiology: the lifelong interplay of genes, lifestyle, and hormones. Breast Cancer Res 6:213–218. https://doi.org/10.1186/bcr921
Stebbing J, Filipovic A, Giamas G (2011) Lemure tyrosine kinase-3 (LMTK3) in cancer evolution. Oncotarget 2:428–429. https://doi.org/10.18632/oncotarget.291
Lewis-Wambi JS, Jordan VC (2006) Treatment of postmenopausal breast cancer with selective estrogen receptor modulators (SERMs). Breast Dis 24:93–105. https://doi.org/10.3233/bd-2006-24108
Leary AA, Dowsett M (2006) Combination therapy with aromatase inhibitors: the next era of breast cancer treatment? Br J Cancer 95:661–666. https://doi.org/10.1038/sj.bjc.6603316
Ali S, Coombes C (2002) Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer 2:101–112. https://doi.org/10.1038/nrc721
Lannigan DA (2003) Estrogen receptor phosphorylation. Steroids 68:1–9. https://doi.org/10.1016/s0039-128x(02)00110-1
Thomas RS, Sarwar N, Phoenix F, Coombes RC, Ali S (2008) Phosphorylation at serines 104 and 106 by Erk1/2 MAPK is important for estrogen receptor-alpha activity. J Mol Endocrinol 40:173–84. https://doi.org/10.1677/jme-07-0165
Rogatsky I, Trowbridge JM, Garabediani MJ (1999) Potentiation of human estrogen receptor a transcriptional activation through phosphorylation of serines 104 and 106 by the cyclin A-CDK2 complex. J Biol Chem 274:22296–22302. https://doi.org/10.1074/jbc.274.32.22296
Zwart W, Griekspoor A, Berno V, Lakeman K, Jalink K, Mancini M, Neefjes J, Michalides R (2007) PKA-induced resistance to tamoxifen is associated with an altered orientation of ERα towards co-activator SRC-1. EMBO J 26:3534–3544. https://doi.org/10.1038/sj.emboj.7601791
Jiang J, Sarwar N, Peston D, Kulinskaya E, Shousha S, Coombes RC, Ali S (2007) Phosphorylation of estrogen receptor-alpha at ser-167 is indicative of longer disease free and overall survival in breast cancer patients. Clin Cancer Res 13:5769–5776. https://doi.org/10.1158/1078-0432.ccr-07-0822
Giamas G, Castellano L, Feng Q, Knippschild U, Jacob J, Thomas RS, Coombes RC, Smith CL, Jiao LR, Stebbing J (2009) CK1 delta modulates the transcriptional activity of ER alpha via AIB1 in an estrogen-dependent manner and regulates ERalpha–AIB1 interactions. Nucleic Acids Res 37:3110–3123. https://doi.org/10.1093/nar/gkp136
Giamas G, Stebbing J, Vorgias CE, Knippschild U (2007) Protein kinas as target for cancer treatment. Pharmacogenomics 8:1005–1016. https://doi.org/10.2217/14622416.8.8.1005
Inoue T, Kon T, Ohkura R, Yamakawa H, Ohara O, Yokota J, Sutoh K (2008) BREK/LMTK3 is a myosin VI-binding protein involved in endosomal membrane trafficking. Genes Cells 13:483–495. https://doi.org/10.1111/j.1365-2443.2008.01184.x
Robinson DR, Wu YM, Lin SF (2000) The protein tyrosine kinase family of the human genome. Oncogene 19:5548–5557. https://doi.org/10.1038/sj.onc.1203957
Giamas G, Filipovic A, Jacob J, Messier W, Zhang H, Yang D, Zhang W, Shifa BA, Photiou A, Tralau-Stewart C, Castellano L, Green AR, Coombes RC, Ellis IO, Ali S, Lenz HJ, Stebbing J (2011) Kinome screening for regulators of the estrogen receptor identifies LMTK3 as a new therapeutic target in breast cancer. Nat Med 17:715–719. https://doi.org/10.1038/nm.2351
Stebbing J, Filipovic A, Ellis IO, Green AR, D’Silva TR, Lenz HJ, Coombes RC, Wang T, Lee SC, Giamas G (2012) LMTK3 expression in breast cancer: association with tumour phenotype and clinical outcome. Breast Cancer Res Treat 132:537–544. https://doi.org/10.1007/s10549-011-1622-z
Stebbing J, Filipovic A, Lit LC, Blighe K, Grothey A, Xu Y, Miki Y, Chow LW (2013) LMTK3 is implicated in endocrine resistance via multiple signaling pathway. Oncogene 32:3371–3380. https://doi.org/10.1038/onc.2012.343
Xu Y, Zhang H, Lit LC, Grothey A, Athanasiadou M, Kiritsi M, Lombardo Y, Frampton AE, Green AR, Ellis IO, Ali S, Lenz HJ, Thanou M, Stebbing J, Giamas G (2014) The kinase LMTK3 promotes invasion in breast cancer through GBR2-mediated induction of integrin β1. Sci Signal 7:330ra58. https://doi.org/10.1126/scisignal.2005170
Xu Y, Zhang H, Giamas G (2014) Targeting lemurs against cancer metastasis. Oncotarget 5:5192–5193. https://doi.org/10.18632/oncotarget.2271
Anbarasu K, Jayanthi S (2014) Structural modeling and molecular dynamics studies on human LMTK3 domain and the mechanism of ATP binding. MolBiosyst 10:1139–1145. https://doi.org/10.1039/c4mb00063c
Anbarasu K, Jayanthi S (2016) Designing and optimization of novel human LMTK3 inhibitors against breast cancer—a computational approach. J Recept Signal Transduct 8:1–9. https://doi.org/10.3109/10799893.2016.1155069
Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. https://doi.org/10.1038/nprot.2010.5
Sarma H, Mattaparthi VSK (2017) Unveiling the transient protein–protein interactions that modulate the activity of estrogen receptor (ER)-α by human lemur tyrosine kinase-3 (LMTK3) domain: an in silico study. Curr Proteom 14:1–8. https://doi.org/10.2174/1570164614666161206164330
Huang D, Zhou T, Lafleur K, Nevado C, Caflisch A (2009) Kinase selectivity potential for inhibitors targeting the ATP binding site: a network analysis. Bioinformatics 26:198–204. https://doi.org/10.1093/bioinformatics/btp650
Stamos J, Sliwkowski MX, Eigenbrot C (2002) Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem 277:46265–46272. https://doi.org/10.1074/jbc.m207135200
Irwin JJ, Shoichet BK, Mysinger MM, Huang N, Colizzi N, Wassam F, Cao Y (2009) Automated docking screen: a feasibility study. J Med Chem 52:5712–5720. https://doi.org/10.1021/jm9006966
Shrivastava AK, Kumar S, Sahu PS, Mahapatra RK (2017) In silico identification and validation of a novel hypothetical protein in Cryptosporidium hominis and virtual screening of inhibitors as therapeutics. Parasitol Res 116:1533–1544. https://doi.org/10.1007/s00436-017-5430-1
Sodero ACR, Dos Santos ACG, MELLO JFE, De Jesus JB, De Souza AM, Rodrigues MIC, Simone SGD, Rodrigues CR, Guedes HLDM (2017) Oligopeptidase B and B2: comparative modelling and virtual screening as searching tools for new antileishmanial compounds. Parasitology 144:536–545. https://doi.org/10.1017/s0031182016002237
Arrigoni A, Bertini L, De Gioia L, Papaleo E (2014) Inhibitors of the Cdc34 acidic loop: a computational investigation integrating molecular dynamics, virtual screening and docking approaches. FEBS open bio 4:473–484. https://doi.org/10.1016/j.fob.2014.04.011
Hou T, Wang J, Li Y, Wang W (2011) Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model 51:69–82. https://doi.org/10.1021/ci100275a
Kollman PA, Massova I, Reyes C, Kuhn B, Huo SH, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 33:889–897. https://doi.org/10.1021/ar000033j
Wang W, Donini O, Reyes CM, Kollman PA (2001) Biomolecular simulations: recent developments in force fields, simulations of enzyme catalysis, protein–ligand, protein–protein, and protein–nucleic acid non-covalent interactions. Annu Rev Biophys Biomol Struct 30:211–243. https://doi.org/10.1146/annurev.biophys.30.1.211
Wang J, Hou T, Xu X (2006) Recent advances in free energy calculations with a combination of molecular mechanics and continuum models. Curr Comput Aided Drug Des 2:287–306. https://doi.org/10.2174/157340906778226454
Morries GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. https://doi.org/10.1002/jcc.21256
Laskowski RA, Swindells MB (2011) LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J Chem Inf Model 51:2778–2786. https://doi.org/10.1021/ci200227u
Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. ZINC: a free tool to discover chemistry for biology. J Chem Inf Model 52:1757–1768. https://doi.org/10.1021/ci3001277
Coleman RG, Sharp KA (2010) Protein pockets: inventory, shape, and comparison. J Chem Inf Model 50:589–603. https://doi.org/10.1021/ci900397t
Lipinski CA (2004) Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol 1:337–341. https://doi.org/10.1016/j.ddtec.2004.11.007
Kaus JW, Pierce LT, Walker RC, McCammont JA (2013) Improving the efficiency of free energy calculations in the amber molecular dynamics package.. J Chem Theory Comput 9:4131–4139. https://doi.org/10.1021/ct400340s
Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Hayik S, Roitberg A, Seabra G, Swails J, Götz AW, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wolf RM, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh MJ, Cui G, Roe DR, Mathews DH, Seetin MG, Salomon-Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA (2012) AMBER 12. University of California, San Francisco
Wang J, Wang W, Kollman PA, Case DA (2006) Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model 25:247–260. https://doi.org/10.1016/j.jmgm.2005.12.005
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general amber force field.. J Comput Chem 25:1157–1174. https://doi.org/10.1002/jcc.20035
Jakalian A, Jack DB, Bayly CI (2002) Fast, efficient generation of high-quality atomic charges. AM1-BCC Model: II. Parameterization and validation. J Comput Chem 23:1623–1641. https://doi.org/10.1002/jcc.10128
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935. https://doi.org/10.1063/1.445869
Berendsen HJC, Postma JRM, van Gunsteren WE, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690. https://doi.org/10.1063/1.448118
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23:327–341. https://doi.org/10.1016/0021-9991(77)90098-5
Daniel RR, Cheatham TE (2013) PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput 9:3084–3095. https://doi.org/10.1021/ct400341p
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14:33–38. https://doi.org/10.1016/0263-7855(96)00018-5
Massova I, Kollman PA (2000) Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspect Drug Discov Des 18:113–135. https://doi.org/10.1023/A:1008763014207
Torrie GM, Valleau JP (1977) Nonphysical sampling distributions in Monte Carlo free energy estimation: umbrella sampling. J Comput Phys 23:187–199. https://doi.org/10.1016/0021-9991(77)90121-8
Kumar S, Rosenberg JM, Bouzida D, Swendsen RH, Kollman PA (1992) The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J Comput Chem 13:1011–1021. https://doi.org/10.1002/jcc.540130812
Souaille M, Roux B (2001) Extension to the weighted histogram analysis method: combining umbrella sampling with free energy calculations. Comput Phys Commun 135:40–57. https://doi.org/10.1016/s0010-4655(00)00215-0
Sun H, Li Y, Li D, Hou T (2013) Insight into crizotinib resistance mechanisms caused by three mutations in ALK tyrosine kinase using free energy calculation approaches. J Chem Inf Model 53:2376–2389. https://doi.org/10.1021/ci400188q
Sun H, Li Y, Tian S, Wanq J, Hou T (2014) P-loop conformation governed crizotinib resistance in G2032R-mutated ROS1 tyrosine kinase: clues from free energy landscape. PLoS Comput Biol 10:e1003729. https://doi.org/10.1371/journal.pcbi.1003729
Sun H, Tian S, Zhou S, Li Y, Li D, Xu L, Shen M, Panand P, Hou T (2015) Revealing the favorable dissociation pathway of type II kinase inhibitors via enhanced sampling simulations and two-end-state calculations. Sci Rep 5:8457. https://doi.org/10.1038/srep08457
Taylor SS, Knighton DR, Zheng J, Sowadski JM, Gibbs CS, Zoller MJ (1993) A template for the protein kinase family. Trends Biochem Sci 18:84–89. https://doi.org/10.1016/0968-0004(93)80001-R
Nolen B, Taylor S, Ghosh G (2004) Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell 15:661–675. https://doi.org/10.1016/j.molcel.2004.08.024
Allam L, Lakhlili W, Tarhda Z, Akachar J, Ghrifi F, Amri HE, Ibrahimi A (2017) Three-dimensional structure prediction of the human LMTK3 catalytic domain in DYG-in conformation. J Biomol Res Ther 6:1000151. https://doi.org/10.4172/2167-7956.1000151
Sarma H, Mattaparthi VSK (2018) Unveiling the transient protein–protein interactions that regulate the activity of human lemur tyrosine kinase-3 (LMTK3) domain by cyclin dependent kinase 5 (CDK5) in breast cancer: an in silico study. Curr Proteom 15:62–70. https://doi.org/10.2174/1570164614666161206164330
Noble MEM, Endicott JA, Johnson LN (2004) Protein kinase inhibitors: insights into drug design from structure. Science 30:1800–1805. https://doi.org/10.1126/science.1095920
Pargellis TL, Churchill L, Cirillo PF, Gilmore T, Graham AG, Grob PM, Hickey ER, Moss N, Pav S, Regan J (2002) Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site. Nat Struct Biol 9:268–272. https://doi.org/10.1038/nsb770
Kwarcinski FE, Brandvold KR, Phadke S, Beleh OM, Johnson TK, Meagher JL, Seeliger MA, Stuckey JA, Soellner MB (2016) Conformation-selective analogues of dasatinib reveal insight into kinase inhibitor binding and selectivity. ACS Chem Biol 11:1296–1304. https://doi.org/10.1021/acschembio.5b01018
Roskoski RJ (2016) Classification of small molecule protein kinase inhibitors based upon the structures of their drug-enzyme complexes. Pharmacol Res 103:26–48. https://doi.org/10.1016/j.phrs.2015.10.021
Suebsuwong C, Pinkas DM, Ray SS, Bufton JC, Dai B, Bullock AN, Degterev A, Cuny GD (2018) Activation loop targeting strategy for design of receptor-interacting protein kinase 2 (RIPK2) inhibitors. Bioorg Med Chem Lett 28:577–583. https://doi.org/10.1016/j.bmcl.2018.01.044
Park JW, Jo WH (2010) Computational design of novel, high-affinity neuraminidase inhibitors for H5N1 avian influenza virus. Eur J Med Chem 45:536–541. https://doi.org/10.1016/j.ejmech.2009.10.040
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2012) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26. https://doi.org/10.1016/S0169-409X(00)00129-0
Acknowledgements
We would like to thank Tezpur University and UGC for the start-up Grant, and DBT funded Bioinformatics Infrastructure facility in the Department of Molecular Biology and Biotechnology at Tezpur University for providing us computational facility to carry out this research work. We would also like to thank Miss Sushmita Pradhan, Research Scholar at Molecular Modelling and Simulation Laboratory, Department of MBBT, Tezpur University for helping us in the preparation of figures.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sarma, H., Mattaparthi, V.S.K. Structure-Based Virtual Screening of High-Affinity ATP-Competitive Inhibitors Against Human Lemur Tyrosine Kinase-3 (LMTK3) Domain: A Novel Therapeutic Target for Breast Cancer. Interdiscip Sci Comput Life Sci 11, 527–541 (2019). https://doi.org/10.1007/s12539-018-0302-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-018-0302-7