Abstract
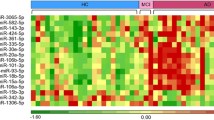
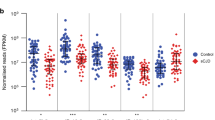
A series of investigations have been performed regarding microRNA (miRNA, miR) of Alzheimer’s disease (AD) patients. However, most of these used microarray with neither validation by PCR nor any follow-up on the biological mechanism implicated by findings. Further, there were rarely any analyses linking clinical phenotype of de novo, drug-naive patients to cellular pathogenic mechanism(s) to date. Microarray screening followed by validation via quantitative PCR (Q-PCR) assays and the relationship between miRNAs and phenotypic indices were evaluated. Additionally, the cellular mechanism of miRNAs through effects of β-site amyloid precursor protein (APP) cleaving enzyme (BACE1) was assessed. We identified 2 specific differentially expressed (DE) miRNAs (miR-339 and miR-425) as potential diagnostic biomarkers for AD and revealed that these DE miRNAs could be involved in modulating the pathogenesis of AD via BACE1 protein inhibition. The findings presented here reveal a detailed snapshot of the profile of peripheral blood mononuclear cells (PBMC) miRNA changes in AD patients, association with clinical phenotype, and potential roles in cellular pathogenesis.




Similar content being viewed by others
References
Duyckaerts C, Delatour B, Potier MC (2009) Classification and basic pathology of Alzheimer disease. Acta Neuropathol 118:5–36. doi:10.1007/s00401-009-0532-1
Hardy J (1997) Amyloid, the presenilins and Alzheimer’s disease. Trends Neurosci 20:154–9
Chen XF, Zhang YW, Xu H, Bu G (2013) Transcriptional regulation and its misregulation in Alzheimer’s disease. Mol Brain 6:44. doi:10.1186/1756-6606-6-44
Cataldo AM, Petanceska S, Peterhoff CM, Terio NB, Epstein CJ, Villar A, Carlson EJ, Staufenbiel M et al (2003) App gene dosage modulates endosomal abnormalities of Alzheimer’s disease in a segmental trisomy 16 mouse model of down syndrome. J Neurosci 23:6788–92
Kim DH, Yeo SH, Park JM, Choi JY, Lee TH, Park SY, Ock MS, Eo J et al (2014) Genetic markers for diagnosis and pathogenesis of Alzheimer’s disease. Gene 545:185–93. doi:10.1016/j.gene.2014.05.031
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–54
Vo NK, Cambronne XA, Goodman RH (2010) MicroRNA pathways in neural development and plasticity. Curr Opin Neurobiol 20:457–65. doi:10.1016/j.conb.2010.04.002
Bilen J, Liu N, Bonini NM (2006) A new role for microRNA pathways: modulation of degeneration induced by pathogenic human disease proteins. Cell Cycle 5:2835–8
Luo H, Wu Q, Ye X, Xiong Y, Zhu J, Xu J, Diao Y, Zhang D et al (2014) Genome-wide analysis of miRNA signature in the APPswe/PS1DeltaE9 mouse model of Alzheimer’s disease. PLoS One 9, e101725. doi:10.1371/journal.pone.0101725
Burgos K, Malenica I, Metpally R, Courtright A, Rakela B, Beach T, Shill H, Adler C et al (2014) Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer’s and Parkinson’s diseases correlate with disease status and features of pathology. PLoS One 9, e94839. doi:10.1371/journal.pone.0094839
Li YY, Cui JG, Hill JM, Bhattacharjee S, Zhao Y, Lukiw WJ (2011) Increased expression of miRNA-146a in Alzheimer’s disease transgenic mouse models. Neurosci Lett 487:94–8. doi:10.1016/j.neulet.2010.09.079
Rodriguez-Ortiz CJ, Baglietto-Vargas D, Martinez-Coria H, LaFerla FM, Kitazawa M (2014) Upregulation of miR-181 decreases c-Fos and SIRT-1 in the hippocampus of 3xTg-AD mice. J Alzheimers Dis 42:1229–38. doi:10.3233/JAD-140204
Tan L, Yu JT, Tan MS, Liu QY, Wang HF, Zhang W, Jiang T (2014) Genome-wide serum microRNA expression profiling identifies serum biomarkers for Alzheimer’s disease. J Alzheimers Dis 40:1017–27. doi:10.3233/JAD-132144
Sun X, Wu Y, Gu M, Zhang Y (2014) miR-342-5p decreases ankyrin G levels in Alzheimer’s disease transgenic mouse models. Cell Rep 6:264–70. doi:10.1016/j.celrep.2013.12.028
Long JM, Ray B, Lahiri DK (2014) MicroRNA-339-5p down-regulates protein expression of beta-site amyloid precursor protein-cleaving enzyme 1 (BACE1) in human primary brain cultures and is reduced in brain tissue specimens of Alzheimer disease subjects. J Biol Chem 289:5184–98. doi:10.1074/jbc.M113.518241
Sala Frigerio C, Lau P, Salta E, Tournoy J, Bossers K, Vandenberghe R, Wallin A, Bjerke M et al (2013) Reduced expression of hsa-miR-27a-3p in CSF of patients with Alzheimer disease. Neurology 81:2103–6. doi:10.1212/01.wnl.0000437306.37850.22
Tan L, Yu JT, Liu QY, Tan MS, Zhang W, Hu N, Wang YL, Sun L et al (2014) Circulating miR-125b as a biomarker of Alzheimer’s disease. J Neurol Sci 336:52–6. doi:10.1016/j.jns.2013.10.002
Leidinger P, Backes C, Deutscher S, Schmitt K, Mueller SC, Frese K, Haas J, Ruprecht K et al (2013) A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol 14:R78. doi:10.1186/gb-2013-14-7-r78
Salazar C, Nagadia R, Pandit P, Cooper-White J, Banerjee N, Dimitrova N, Coman WB, Punyadeera C (2014) A novel saliva-based microRNA biomarker panel to detect head and neck cancers. Cell Oncol (Dordr) 37:331–8. doi:10.1007/s13402-014-0188-2
Boeckel JN, Thome CE, Leistner D, Zeiher AM, Fichtlscherer S, Dimmeler S (2013) Heparin selectively affects the quantification of microRNAs in human blood samples. Clin Chem 59:1125–7. doi:10.1373/clinchem.2012.199505
Kroh EM, Parkin RK, Mitchell PS, Tewari M (2010) Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 50:298–301. doi:10.1016/j.ymeth.2010.01.032
Dong Y, Yu J, Ng SS (2014) MicroRNA dysregulation as a prognostic biomarker in colorectal cancer. Cancer Manag Res 6:405–22. doi:10.2147/CMAR.S35164
Zhu W, Kan X (2014) Neural network cascade optimizes microRNA biomarker selection for nasopharyngeal cancer prognosis. PLoS One 9, e110537. doi:10.1371/journal.pone.0110537
Lukiw WJ (2007) Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport 18:297–300. doi:10.1097/WNR.0b013e3280148e8b
Villa C, Fenoglio C, De Riz M, Clerici F, Marcone A, Benussi L, Ghidoni R, Gallone S et al (2011) Role of hnRNP-A1 and miR-590-3p in neuronal death: genetics and expression analysis in patients with Alzheimer disease and frontotemporal lobar degeneration. Rejuvenation Res 14:275–81. doi:10.1089/rej.2010.1123
Tierney MC, Fisher RH, Lewis AJ, Zorzitto ML, Snow WG, Reid DW, Nieuwstraten P (1988) The NINCDS-ADRDA Work Group criteria for the clinical diagnosis of probable Alzheimer’s disease: a clinicopathologic study of 57 cases. Neurology 38:359–64
Zivelin A, Rosenberg N, Peretz H, Amit Y, Kornbrot N, Seligsohn U (1997) Improved method for genotyping apolipoprotein E polymorphisms by a PCR-based assay simultaneously utilizing two distinct restriction enzymes. Clin Chem 43:1657–9
Maes OC, Xu S, Yu B, Chertkow HM, Wang E, Schipper HM (2007) Transcriptional profiling of Alzheimer blood mononuclear cells by microarray. Neurobiol Aging 28:1795–809. doi:10.1016/j.neurobiolaging.2006.08.004
Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB (2003) Prediction of mammalian microRNA targets. Cell 115:787–98
Zhang B, Kirov S, Snoddy J (2005) WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 33:W741–8. doi:10.1093/nar/gki475
Schipper HM, Maes OC, Chertkow HM, Wang E (2007) MicroRNA expression in Alzheimer blood mononuclear cells. Gene Regul Syst Bio 1:263–74
Deng Y, Ding Y, Hou D (2014) Research status of the regulation of miRNA on BACE1. Int J Neurosci 124:474–7. doi:10.3109/00207454.2013.858249
Reitz C (2012) The role of intracellular trafficking and the VPS10d receptors in Alzheimer’s disease. Future Neurol 7:423–431. doi:10.2217/fnl.12.31
Pascual-Lucas M, Viana da Silva S, Di Scala M, Garcia-Barroso C, Gonzalez-Aseguinolaza G, Mulle C, Alberini CM, Cuadrado-Tejedor M et al (2014) Insulin-like growth factor 2 reverses memory and synaptic deficits in APP transgenic mice. EMBO Mol Med 6:1246–62. doi:10.15252/emmm.201404228
Wu ZS, Wu Q, Wang CQ, Wang XN, Wang Y, Zhao JJ, Mao SS, Zhang GH et al (2010) MiR-339-5p inhibits breast cancer cell migration and invasion in vitro and may be a potential biomarker for breast cancer prognosis. BMC Cancer 10:542. doi:10.1186/1471-2407-10-542
Zhou C, Liu G, Wang L, Lu Y, Yuan L, Zheng L, Chen F, Peng F et al (2013) MiR-339-5p regulates the growth, colony formation and metastasis of colorectal cancer cells by targeting PRL-1. PLoS One 8, e63142. doi:10.1371/journal.pone.0063142
Liu J, Li T, Zhang N, Yang X, Wang Z, Ma J, Gu X, Fan Y et al (2014) MiR-425 up-regulation induced by interleukin-1beta promotes the proliferation of gastric cancer cell AGS. Zhonghua Yi Xue Za Zhi 94:1889–93
Acknowledgments
The authors are grateful to all of the subjects who kindly agreed to participate in this study. We also thank Dr. Lian Li at the Department of Pharmacology, Emory University School of Medicine, for critical reading and comments on this manuscript. This study was supported by the National Basic Research Development Program of China (No. 2010CB945200, 2011CB504104), and the National Natural Science Foundation of China (No. 81371218, 81001426, 81070958, 81171027, 81200842, 81573401,91332107).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no competing interests.
Ethical Approval
The study was approved by the Research Ethics Committee of Ruijin Hospital affiliated with Shanghai Jiao Tong University School of Medicine in China.
Informed Consent
Written informed consent was obtained from each participant.
Additional information
Ru-Jing Ren and Yong-Fang Zhang contributed equally to this work.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
Demographics of subjects in the study (DOC 34 kb)
ESM 2
Real-time PCR Primer used in the study (DOC 32 kb)
ESM 3
The correlation between DE miRNAs and clinical phenotypes among AD group (n=30) (DOC 29 kb)
ESM 4
Differentially Expressed miRNAs (Pass Volcano Plot) (XLS 84 kb)
ESM 5
Targeted gene prediction (XLS 48 kb)
Figure S1
MiRNAs-gene network in the present study (JPEG 71 kb)
Rights and permissions
About this article
Cite this article
Ren, RJ., Zhang, YF., Dammer, E.B. et al. Peripheral Blood MicroRNA Expression Profiles in Alzheimer’s Disease: Screening, Validation, Association with Clinical Phenotype and Implications for Molecular Mechanism. Mol Neurobiol 53, 5772–5781 (2016). https://doi.org/10.1007/s12035-015-9484-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-015-9484-8