Abstract
Background
Glioblastoma multiforme (GBM) is the most prevalent and malignant intracranial tumor with significant features of dismal prognosis and limited therapeutic solutions. Consequently, the present studies are committed to exploring potential biomarkers through bioinformatics analysis, which may serve as valuable prognostic predictors or novel therapeutic targets and provide new insights into the pathogenesis of GBM.
Methods
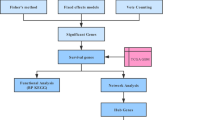
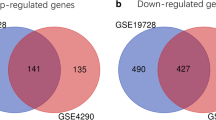
We filtered overlapping differentially expressed genes (DEGs) based on expression profilings from three GBM microarray datasets (GSE116520, GSE4290 and GSE68848) and combined RNA sequencing data from The Cancer Genome Atlas and the Genotype-Tissue Expression databases. Hub genes were prioritized from DEGs after performing protein–protein interaction (PPI) network analysis and weighted gene co-expression network analysis (WGCNA). This was followed by survival analysis to identify potential biomarkers among hub genes. Ultimately, the distributions of gene expressions, genetic alterations, upstream regulatory mechanisms and enrichments of gene functions of the identified biomarkers were analysed on public databases. QRT-PCR, immunohistochemical staining and western blotting was also used to confirm the gene expression patterns in GBM and normal brain tissues. CCK-8 assay clarified the effects of the genes on GBM cells.
Results
A total of 322 common DEGs were determined and nine genes were subsequently considered as hub genes by the combination of PPI network analysis and WGCNA. Only SLC12A5 had prognostic significance, which was deficient in GBM whereas especially enriched in normal neural tissues. SLC12A5 overexpression would inhibit cell proliferation of U251MG. Genetic alterations of SLC12A5 were rarely seen in GBM patients, and there was no apparent association existed between SLC12A5 expression and DNA methylation. SLC12A5 was prominently involved in ion transport, synapse and neurotransmitter.
Conclusion
SLC12A5 shows promise to function as a novel effective biomarker for GBM and deserves further systematic research.






Similar content being viewed by others
Data availability
The web links of the related public datasets or databases are as follows: GSE116520: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE116520; GSE4290: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4290; GSE68848: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE68848; GEPIA: http://gepia.cancer-pku.cn/; STRING: http://string.embl.de/; CGGA: http://cgga.org.cn/; HPA: http://www.proteinatlas.org/; cBioportal: https://www.cbioportal.org/; NCBI: http://www.ncbi.nlm.nih.gov; JASPAR: https://jaspar.genereg.net/; Metascape: http://metascape.org/; GSEA: http://www.broad.mit.edu/gsea/; GeneMANIA: http://genemania.org/. All the accesses above are open. Other data are available upon request.
Abbreviations
- GBM:
-
Glioblastoma multiforme
- CALD1:
-
Caldesmon 1
- TREM1:
-
Triggering receptor expressed on myeloid cells 1
- TMZ:
-
Temozolomide
- PLK2:
-
Polo like kinase 2
- GEO:
-
Gene expression omnibus
- DEG:
-
Differentially expressed gene
- TCGA:
-
The cancer genome atlas
- GTEx:
-
Genotype-tissue expression
- PPI:
-
Protein–protein interaction
- WGCNA:
-
Weighted gene co-expression network analysis
- CGGA:
-
Chinese glioma genome atlas
- FC:
-
Fold change
- GEPIA:
-
Gene expression profiling interactive analysis
- RNA:
-
Ribonucleic acid
- STRING:
-
Search tool for the retrieval of interaction genes
- MCODE:
-
Molecular complex detection
- OS:
-
Overall survival
- cDNA:
-
Complementary deoxyribonucleic acid
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- IHC:
-
Immunohistochemical
- CCK-8:
-
Cell counting Kit-8
- HPA:
-
Human protein atlas
- cBioportal:
-
CBio cancer genomics portal
- GO:
-
Gene ontology
- BP:
-
Biological process
- CC:
-
Cellular component
- MF:
-
Molecular function
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- GSEA:
-
Gene set enrichment analysis
- HR:
-
Hazard ratio
- LGG:
-
Lower grade glioma
- GPCR:
-
G Protein-coupled receptor
- SLC12A5:
-
Solute carrier family 12 member 5
- KCC2:
-
K( +)-Cl(−) cotransporter-2
- GABA:
-
Gamma-aminobutyric acid
- MECP2:
-
Methyl-CpG binding protein 2
- REST:
-
RE1 silencing transcription factor
- EGR4:
-
Early growth response 4
- SOX18:
-
SRY-box transcription factor 18
- PATZ1:
-
POZ/BTB and AT hook containing zinc finger 1
- SP2:
-
Specific protein 2
- ARNT2:
-
Aryl hydrocarbon receptor nuclear translocator 2
- USF2:
-
Upstream stimulatory factor 2
- UTR:
-
Untranslated region
- TWIST1:
-
Twist family bHLH transcription factor 1
References
Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM, Reifenberger G, Soffietti R, von Deimling A, Ellison DW (2021) The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol 23(8):1231–1251. https://doi.org/10.1093/neuonc/noab106
Stupp R, Hegi ME, Mason WP, Bent M, Mirimanoff RO (2009) Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 10(5):434–435. https://doi.org/10.1016/S1470-2045(09)70025-7
Hu LS, Hawkins-Daarud A, Wang L, Li J, Swanson KR (2020) Imaging of intratumoral heterogeneity in high-grade glioma. Cancer Lett 477:97–106. https://doi.org/10.1016/j.canlet.2020.02.025
Yang K, Wu Z, Zhang H, Zhang N, Wu W, Wang Z, Dai Z, Zhang X, Zhang L, Peng Y, Ye W, Zeng W, Liu Z, Cheng Q (2022) Glioma targeted therapy: insight into future of molecular approaches. Mol Cancer 21(1):39. https://doi.org/10.1186/s12943-022-01513-z
Cheng Q, Tang A, Wang Z, Fang N, Zhang Z, Zhang L, Li C, Zeng Y (2021) CALD1 modulates gliomas progression via facilitating tumor angiogenesis. Cancers. https://doi.org/10.3390/cancers13112705
Kong Y, Feng Z, Zhang Y, Liu X, Ma Y, Zhao Z, Huang B, Chen A, Zhang D, Thorsen F, Wang J, Yang N, Li X (2020) Identification of immune-related genes contributing to the development of glioblastoma using weighted gene co-expression network analysis. Front Immunol 11:1281. https://doi.org/10.3389/fimmu.2020.01281
Alafate W, Xu D, Wu W, Xiang J, Ma X, Xie W, Bai X, Wang M, Wang J (2020) Loss of PLK2 induces acquired resistance to temozolomide in GBM via activation of notch signaling. J Exp Clin Cancer Res 39(1):239. https://doi.org/10.1186/s13046-020-01750-4
Barrett T, Wilhite S, Ledoux P, Evangelista C, Kim I, Tomashevsky M, Marshall K, Phillippy K, Sherman P, Holko M, Yefanov A, Lee H, Zhang N, Robertson C, Serova N, Davis S, Soboleva A (2013) NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res 41:D991-995. https://doi.org/10.1093/nar/gks1193
Kruthika B, Jain R, Arivazhagan A, Bharath R, Yasha T, Kondaiah P, Santosh V (2019) Transcriptome profiling reveals PDZ binding kinase as a novel biomarker in peritumoral brain zone of glioblastoma. J Neurooncol 141(2):315–325. https://doi.org/10.1007/s11060-018-03051-5
Sun L, Hui A, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey R, Rosenblum M, Mikkelsen T, Fine H (2006) Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell 9(4):287–300. https://doi.org/10.1016/j.ccr.2006.03.003
Madhavan S, Zenklusen J, Kotliarov Y, Sahni H, Fine H, Buetow K (2009) Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res MCR 7(2):157–167. https://doi.org/10.1158/1541-7786.Mcr-08-0435
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45:W98–W102. https://doi.org/10.1093/nar/gkx247
Szklarczyk D, Gable A, Nastou K, Lyon D, Kirsch R, Pyysalo S, Doncheva N, Legeay M, Fang T, Bork P, Jensen L, von Mering C (2021) The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 49:D605–D612. https://doi.org/10.1093/nar/gkaa1074
Ge S, Son E, Yao R (2018) iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinfo 19(1):534. https://doi.org/10.1186/s12859-018-2486-6
Zhao Z, Zhang K, Wang Q, Li G, Zeng F, Zhang Y, Wu F, Chai R, Wang Z, Zhang C, Zhang W, Bao Z, Jiang T (2021) Chinese glioma genome atlas (CGGA): a comprehensive resource with functional genomic data from Chinese glioma patients. Genom Proteom Bioinfo. https://doi.org/10.1016/j.gpb.2020.10.005
Zhao Z, Meng F, Wang W, Wang Z, Zhang C, Jiang T (2017) Comprehensive RNA-seq transcriptomic profiling in the malignant progression of gliomas. Scientific Data 4:170024. https://doi.org/10.1038/sdata.2017.24
Wang Y, Qian T, You G, Peng X, Chen C, You Y, Yao K, Wu C, Ma J, Sha Z, Wang S, Jiang T (2015) Localizing seizure-susceptible brain regions associated with low-grade gliomas using voxel-based lesion-symptom mapping. Neuro Oncol 17(2):282–288. https://doi.org/10.1093/neuonc/nou130
Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, Sanli K, von Feilitzen K, Oksvold P, Lundberg E, Hober S, Nilsson P, Mattsson J, Schwenk J, Brunnström H, Glimelius B, Sjöblom T, Edqvist P, Djureinovic D, Micke P, Lindskog C, Mardinoglu A, Ponten F (2017) A pathology atlas of the human cancer transcriptome. Science. https://doi.org/10.1126/science.aan2507
Cerami E, Gao J, Dogrusoz U, Gross B, Sumer S, Aksoy B, Jacobsen A, Byrne C, Heuer M, Larsson E, Antipin Y, Reva B, Goldberg A, Sander C, Schultz N (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2(5):401–404. https://doi.org/10.1158/2159-8290.Cd-12-0095
Gao J, Aksoy B, Dogrusoz U, Dresdner G, Gross B, Sumer S, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. https://doi.org/10.1126/scisignal.2004088
Hu H, Miao Y, Jia L, Yu Q, Zhang Q, Guo A (2019) AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res 47:D33–D38. https://doi.org/10.1093/nar/gky822
Gene Ontology C (2021) The gene ontology resource: enriching a gold mine. Nucleic Acids Res 49(D1):D325–D334. https://doi.org/10.1093/nar/gkaa1113
Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, Sidiropoulos K, Cook J, Gillespie M, Haw R, Loney F, May B, Milacic M, Rothfels K, Sevilla C, Shamovsky V, Shorser S, Varusai T, Weiser J, Wu G, Stein L, Hermjakob H, D’Eustachio P (2020) The reactome pathway knowledgebase. Nucleic Acids Res 48:D498–D503. https://doi.org/10.1093/nar/gkz1031
Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M (1999) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 27(1):29–34. https://doi.org/10.1093/nar/27.1.29
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi A, Tanaseichuk O, Benner C, Chanda S (2019) Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10(1):1523. https://doi.org/10.1038/s41467-019-09234-6
Subramanian A, Tamayo P, Mootha V, Mukherjee S, Ebert B, Gillette M, Paulovich A, Pomeroy S, Golub T, Lander E, Mesirov J (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102(43):15545–15550. https://doi.org/10.1073/pnas.0506580102
Warde-Farley D, Donaldson S, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes C, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader G, Morris Q (2010) The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38:W214-220. https://doi.org/10.1093/nar/gkq537
Sa J, Chang N, Lee H, Cho H, Ceccarelli M, Cerulo L, Yin J, Kim S, Caruso F, Lee M, Kim D, Oh Y, Lee Y, Her N, Min B, Kim H, Jeong D, Kim H, Kim H, Chung S, Woo H, Lee J, Kong D, Seol H, Lee J, Kim J, Park W, Wang Q, Sulman E, Heimberger A, Lim M, Park J, Iavarone A, Verhaak R, Nam D (2020) Transcriptional regulatory networks of tumor-associated macrophages that drive malignancy in mesenchymal glioblastoma. Genome Biol 21(1):216. https://doi.org/10.1186/s13059-020-02140-x
Payne J, Stevenson T, Donaldson L (1996) Molecular characterization of a putative K-Cl cotransporter in rat brain. A neuronal-specific isoform. J Biol Chem 271(27):16245–16252. https://doi.org/10.1074/jbc.271.27.16245
Liu J, Dai Y, Li X, Cao K, Xie D, Tong Z, Long Z, Xiao H, Chen M, Ye Y, Liu B, Tan J, Tang J, Xu Z, Gan Y, Zhou Y, Deng F, He L (2017) Solute carrier family 12 member 5 promotes tumor invasion/metastasis of bladder urothelial carcinoma by enhancing NF-κB/MMP-7 signaling pathway. Cell Death Dis 8(3):e2691. https://doi.org/10.1038/cddis.2017.118
Xu L, Li X, Cai M, Chen J, Li X, Wu W, Kang W, Tong J, To K, Guan X, Sung J, Chan F, Yu J (2016) Increased expression of Solute carrier family 12 member 5 via gene amplification contributes to tumour progression and metastasis and associates with poor survival in colorectal cancer. Gut 65(4):635–646. https://doi.org/10.1136/gutjnl-2014-308257
Wei W, Akerman C, Newey S, Pan J, Clinch N, Jacob Y, Shen M, Wilkins R, Ellory J (2011) The potassium-chloride cotransporter 2 promotes cervical cancer cell migration and invasion by an ion transport-independent mechanism. J Physiol 589:5349–5359. https://doi.org/10.1113/jphysiol.2011.214635
Campbell S, Robel S, Cuddapah V, Robert S, Buckingham S, Kahle K, Sontheimer H (2015) GABAergic disinhibition and impaired KCC2 cotransporter activity underlie tumor-associated epilepsy. Glia 63(1):23–36. https://doi.org/10.1002/glia.22730
Pallud J, Le Van QM, Bielle F, Pellegrino C, Varlet P, Cresto N, Baulac M, Duyckaerts C, Kourdougli N, Chazal G, Devaux B, Rivera C, Miles R, Capelle L, Huberfeld G (2014) Cortical GABAergic excitation contributes to epileptic activities around human glioma. Sci Trans Med 6(244):244289. https://doi.org/10.1126/scitranslmed.3008065
Conti L, Palma E, Roseti C, Lauro C, Cipriani R, de Groot M, Aronica E, Limatola C (2011) Anomalous levels of Cl- transporters cause a decrease of GABAergic inhibition in human peritumoral epileptic cortex. Epilepsia 52(9):1635–1644. https://doi.org/10.1111/j.1528-1167.2011.03111.x
Venkatesh H, Morishita W, Geraghty A, Silverbush D, Gillespie S, Arzt M, Tam L, Espenel C, Ponnuswami A, Ni L, Woo P, Taylor K, Agarwal A, Regev A, Brang D, Vogel H, Hervey-Jumper S, Bergles D, Suvà M, Malenka R, Monje M (2019) Electrical and synaptic integration of glioma into neural circuits. Nature 573(7775):539–545. https://doi.org/10.1038/s41586-019-1563-y
Yeo M, Berglund K, Augustine G, Liedtke W (2009) Novel repression of Kcc2 transcription by REST-RE-1 controls developmental switch in neuronal chloride. J Neurosci 29(46):14652–14662. https://doi.org/10.1523/jneurosci.2934-09.2009
Tang X, Kim J, Zhou L, Wengert E, Zhang L, Wu Z, Carromeu C, Muotri A, Marchetto M, Gage F, Chen G (2016) KCC2 rescues functional deficits in human neurons derived from patients with Rett syndrome. Proc Natl Acad Sci USA 113(3):751–756. https://doi.org/10.1073/pnas.1524013113
Uvarov P, Ludwig A, Markkanen M, Rivera C, Airaksinen M (2006) Upregulation of the neuron-specific K+/Cl- cotransporter expression by transcription factor early growth response 4. J Neurosci 26(52):13463–13473. https://doi.org/10.1523/jneurosci.4731-06.2006
Wang L, Zhang Q, Wu P, Xiang W, Xie D, Wang N, Deng M, Cao K, Zeng H, Xu Z, Liu X, He L, Long Z, Tan J, Wang J, Liu B, Liu J (2020) SLC12A5 interacts and enhances SOX18 activity to promote bladder urothelial carcinoma progression via upregulating MMP7. Cancer Sci 111(7):2349–2360. https://doi.org/10.1111/cas.14502
Passariello A, Errico ME, Donofrio V, Maestrini M, Zerbato A, Cerchia L, Capasso M, Capasso M, Fedele M (2019) PATZ1 is overexpressed in pediatric glial tumors and correlates with worse event-free survival in high-grade gliomas. Cancers (Basel). https://doi.org/10.3390/cancers11101537
Yu S, Ruan X, Liu X, Zhang F, Wang D, Liu Y, Yang C, Shao L, Liu Q, Zhu L, Lin Y, Xue Y (2021) HNRNPD interacts with ZHX2 regulating the vasculogenic mimicry formation of glioma cells via linc00707/miR-651-3p/SP2 axis. Cell Death Dis 12(2):153. https://doi.org/10.1038/s41419-021-03432-1
Bogeas A, Morvan-Dubois G, El-Habr EA, Lejeune FX, Defrance M, Narayanan A, Kuranda K, Burel-Vandenbos F, Sayd S, Delaunay V, Dubois LG, Parrinello H, Rialle S, Fabrega S, Idbaih A, Haiech J, Bieche I, Virolle T, Goodhardt M, Chneiweiss H, Junier MP (2018) Changes in chromatin state reveal ARNT2 at a node of a tumorigenic transcription factor signature driving glioblastoma cell aggressiveness. Acta Neuropathol 135(2):267–283. https://doi.org/10.1007/s00401-017-1783-x
Markkanen M, Uvarov P, Airaksinen MS (2008) Role of upstream stimulating factors in the transcriptional regulation of the neuron-specific K-Cl cotransporter KCC2. Brain Res 1236:8–15. https://doi.org/10.1016/j.brainres.2008.08.007
He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5(8):631–631
Xia W, Mao Q, Chen B, Wang L, Ma W, Liang Y, Zhang T, Dong G, Xu L, Jiang F (2019) The TWIST1-centered competing endogenous RNA network promotes proliferation, invasion, and migration of lung adenocarcinoma. Oncogenesis 8(11):62. https://doi.org/10.1038/s41389-019-0167-6
Bagla S, Cukovic D, Asano E, Sood S, Luat A, Chugani H, Chugani D, Dombkowski A (2018) A distinct microRNA expression profile is associated with α[C]-methyl-L-tryptophan (AMT) PET uptake in epileptogenic cortical tubers resected from patients with tuberous sclerosis complex. Neurobiol Dis 109:76–87. https://doi.org/10.1016/j.nbd.2017.10.004
Hazra R, Utama R, Naik P, Dobin A, Spector DL (2023) Identification of glioblastoma stem cell-associated lncRNAs using single-cell RNA-sequencing datasets. bioRxiv. https://doi.org/10.1101/2023.01.20.524887
Lei Y, Tang R, Xu J, Wang W, Zhang B, Liu J, Yu X, Shi S (2021) Applications of single-cell sequencing in cancer research: progress and perspectives. J Hematol Oncol 14(1):91. https://doi.org/10.1186/s13045-021-01105-2
Suva ML, Tirosh I (2019) Single-cell RNA sequencing in cancer: lessons learned and emerging challenges. Mol Cell 75(1):7–12. https://doi.org/10.1016/j.molcel.2019.05.003
Acknowledgements
The authors are grateful to Ting Tang and Hui Liang for their assistance in clinical specimen collection, but they may not qualify for authorship, which have been acknowledged with their permission.
Funding
This research was financially supported by the National Natural Science Foundation of China (No.81672503 and No.81702484).
Author information
Authors and Affiliations
Contributions
HW and JC conceived and designed the study. JC, CD and MF carried out experiments and took part in the data collection and analysis. JC wrote the original manuscript. HW reviewed and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interest
The authors declare that they have no competing interests.
Ethical approval
Our study complied with the principles of the Declaration of Helsinki and was approved by the Ethics Committee of Jinling Hospital (No.2017NZGKJ-086).
Informed consent
Written informed consents were sought from patients or related family members prior to specimen collection.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, J., Wang, H., Deng, C. et al. SLC12A5 as a novel potential biomarker of glioblastoma multiforme. Mol Biol Rep 50, 4285–4299 (2023). https://doi.org/10.1007/s11033-023-08371-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08371-y