Abstract
Geminiviridae comprises the largest family of plant viruses which causes severe crop losses in India. The highest pungency chilli Bhut-Jolokia or ghost pepper (Capsicum chinense Jaqc.) hails from North-East region of India and is used in many dishes to add flavors and also for its medicinal value. However, this chilli variety is also affected by viruses leading to crop and economic losses. The present study reports the identification of begomoviruses in the infected chilli Bhut-Jolokia leaf samples collected from eight different places of North-East region (Manipur) of India. The infected leaf samples were screened for the presence of viral genome by rolling circle amplification (RCA) followed by PCR using degenerate primer pairs. The subsequent analyses using restriction fragment length polymorphism and sequencing revealed the presence of Cotton leaf curl Multan virus (CLCuMuV), and Tomato leaf curl Patna betasatellite (ToLCPaB). The findings focus on the phylogenetic relatedness, probable recombinational hot-spots and evolutionary divergence of the viral DNA sequences with the current reported begomoviral genome. To the best of our knowledge, this is the first report showing the presence of CLCuMuV, and associated non-cognate ToLCPaB with leaf curl disease of Bhut-Jolokia chillies. The study reveals potential recombination sites on both viral genome and betsatellite which, during the course of evolution, may have aided the virus to progress and successfully establish infection in chilli plants. Taken together, our results suggest a possible spread of CLCuMuV to the hitherto non-host crop in the North-East region of India.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Capsicum belongs to the family Solanaceae and comprises more than 200 species. Capsicum annum L., Capsicum chinense Jacq., Capsicum frutescens L., Capsicum pubescens L. and Capsicum baccatcm L. are the most frequently cultivated species. Mexico, America, Australia, Britain, Bangladesh, Sri Lanka and India are the major producers of chilli. Bhut-Jolokia (C. chinense Jacq.), is substantially grown in North -East area of India, specifically in Assam, Nagaland and Manipur. In 2007, Bhut-Jolokia made its way to the Guinness World Records as the world’s hottest chilli with 1,001,304 Scoville heat units (SHU’s) [1]. It is commonly known as Bhut-Jolokia (Ghost Chilli) or Bih Jolokia (Poison chilli) in Assam, U-morok (Tree Chilli) in Manipur and Naga-Jolokia in Nagaland. Random amplification of polymorphic DNA (RAPD) marker analysis revealed Bhut-Jolokia to be an inter specific hybrid of C. chinense and C. frutescens [1]. Although, based on phenotype, molecular similarity of internal transcribed spacer (ITS) regions and proteomics evaluation, it has been designated as a distinct species (C. assamicum) [2]. Bhut-Jolokia is used in the form of spice and also known to possess great medicinal properties [3]. However, its production is seriously affected by viral diseases primarily caused by begomoviruses. Plant begomoviruses are the largest dicot infecting genera of the family Geminiviridae and are extensively transmitted by white fly (Bemisia tabaci Genn.). Since viruses are obligatory parasites, the need of host and vector become indispensable for their replication and transmission. The family Geminiviridae is classified into nine genera on the basis of genome structure, mode of spread and varied hosts, of which, begomoviruses are the largest which affect food crops like tomato, potato, chilli, brinjal, radish etc. Chilli leaf curl disease is the most commonly found infection which causes leaf curl in the early stages leading to stunted growth, abscission of flower buds and obstruction of pollen development. The viral infection further enhances the infestation of pests such as thrips and mites causing complete loss of crops [4]. More than twenty species of geminiviruses are identified to be associated with leaf curl disease of chilli/pepper. These include Chilli leaf curl virus (ChiLCV), Chilli leaf curl Palampur virus (ChiLCPV), Tomato leaf curl New Delhi virus (ToLCNDV), Tomato leaf curl Joydebpur virus (ToLCJoV), Chilli leaf curl Vellanad virus (ChiLCVeV) etc. [5,6,7]. The typical symptoms for leaf curl disease are upward and downward leaf curling, puckering of leaves, leaf rolling, yellowing of veins and stunted growth. Association of betasatellites further aggravates disease development through interference with host defense pathways and cause severe phenotypic abnormalities resulting in complete yield loss in several economically important plants [8,9,10,11].
The present paper reports two full-length DNA-A genome of cotton leaf curl multan virus (CLCuMuV) from infected chilli cv. Bhut-Jolokia plants collected from eight different places of Manipur. Also, the screening of viral population led to the isolation of associated Tomato leaf curl Patna betasatellite (ToLCuPaB) with CLCuMuV. The phylogenetic analysis of cloned begomovirus molecules signified their relatedness to CLCuMuV and ToLCuPaB. Further analysis of recombination breakpoints of the begomovirus genome and the per cent GC content of the genome linked with the putative recombination site, revealed the probable pattern of genetic variation occurred during the course of evolution. To the best of our knowledge, this is the first report of association of CLCuMuV from infected Bhut-Jolokia plants marking the spread of the virus to a new host in the North-East region of India.
Materials and methods
Field survey and sample collection
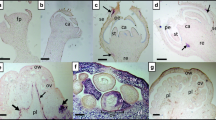
Chilli “Bhut-Jolokia” growing fields were surveyed during the year 2017 at different places viz., Thongju, Heingang, Lamphel, Kiyamgei, Khurai, Koirengei and Andro of Manipur state, India (Fig. 1A). Infected leaf samples were collected based on their symptoms of leaf curling, leaf puckering, and leaf thickening, yellowing of veins and curling of stem (Fig. 1B). The samples were stored at − 80 °C until processed for the detection and cloning of viral genomes.
Field locations and sample collection. A Map displaying geographical regions of chilli var. Bhutjolokia fields used for sample collection at Manipur, (a) Thongju (b) Heingang (c) Lamphel (d) Kiyamgei (e) Khurai (f) Heingang I (g) Koirengei and (h) Andro. B Pictorial representation of infected (I, II, III, IV) and healthy plants of Bhut-Jolokia (V)
Total DNA extraction and rolling circle amplification (RCA)
Total DNA extraction was carried out using the CTAB method [12, 13], and the integrity and quantity were checked using agarose gel electrophoresis and spectrophotometer (Thermo Scientific, USA) respectively. The isolated total DNA samples were then subjected to Rolling Circle Amplification (RCA) using ϕ29 DNA polymerase (Thermo Scientific, USA) to specifically enrich the begomoviral DNA and associated satellite components. The augmented RCA products were further used for individual polymerase chain reactions (PCR) using Phusion high-fidelity DNA polymerase (Thermo Scientific, USA) for amplification of DNA-A, DNA-B, alphasatellite and betasatellite molecules using degenerate primer pairs (available in Molecular Virology Lab, JNU) (Supplementary Table S1).
Cloning of viral genomes
PCR amplified products were purified through the gel-extraction kit (Gene JET™ Gel Extraction Kit, Thermo Scientific, USA) and ligated into blunt-end cloning vector pJET1.2/blunt (Thermo Scientific, USA). The ligated product was used to transform E. coli strain DH5-α competent cells. Transformed clones were randomly selected from each transformation experiment and grown in liquid Luria–Bertani (LB) medium and used for plasmid isolation using the alkaline-lysis method [14]. The plasmids were subjected to restriction digestion with EcoRV for DNA-A clones and KpnI for betasatellite clones.
Restriction fragment length polymorphism (RFLP) and sequencing of viral genome
The positive viral clones were further digested with various restriction enzymes to analyse the pattern for any variation in the viral molecules. The clones with DNA-A molecule were screened using BamHI, PstI, EcoRI and HindIII enzymes whereas clones with betasatellite molecule with EcoRI and PstI (GeNei, India; Thermo Scientific, USA). Up to 24 clones were assessed from each sample to study the polymorphism and the digested products were resolved on 1.2% agarose gel. Since, the profile obtained after restriction analysis showed a similar pattern, one representative clone from each sample was given for sequencing by Sanger’s sequencing at commercial DNA sequencing facility, University of Delhi South Campus, India.
Sequence analysis
The sequence chromatograms obtained were manually scanned and assembled based on the overlapping sequences to assemble the complete viral genome. The reconstructed viral sequences were analysed using NCBI ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/) to examine the coding regions. Sequences were then subjected to standard nucleotide BLAST (blastn) to find the homologous sequences submitted in the GenBank. Sequence datasets were then aligned using multiple sequence comparison by log-expectation algorithm (MUSCLE) followed by molecular evolutionary analysis by Molecular Evolutionary Genetic Analysis tool (MEGA-X) [15].
Phylogenetic relatedness of the sequences obtained was analyzed using the neighbor-joining (NJ) method with 1000 bootstrap values. Based on the earlier literature, the genome sequences of begomoviruses known to infect chilli, cotton, okra and hollyhock plants were selected for the analysis (Supplementary Table S2). Per cent nucleotide identity-based heat maps for both DNA-A and associated betasatellite were generated using sequence demarcation tool (SDT), version1.2 [16].
As geminiviruses are well known for their recombination potential, the sequences were examined for possible recombination events and breakpoints using an algorithm set comprising RDP, GENECONV, Bootscan/Recscan, MaxChi, Chimaera, SiScan and T-3Seq assimilated in recombination detection program (RDP) version 4.97 [17]. Further, the begomoviral genome was analyzed for its GC content using per cent GC-plot graph generated through Artemis DNA plotter analysis tool v18.1.0. (http://www.sanger.ac.uk/Software/Artemis) [18]. Since, both isolated DNA-A molecules were similar at genome sequence level, one of the DNA-A sequences (GenBank accession no. MT886450) was used for further analysis.
The evolutionary divergence between the sequences was calculated by maximum composite likelihood (MCL) model, by using the variance estimation method with 1000 bootstrap values, which include both transition and transversion substitution methods available in MEGA-X tool.
Results
Cloning and sequencing of begomoviral DNA and associated satellite molecule
The viral genome and its associated satellite molecules were PCR amplified post RCA-mediated viral ssDNA enrichment. The desired bands of size ≈ 2.8 and ≈ 1.4 kb were obtained using degenerate primer pair designed for DNA-A and betasatellite, respectively (Supplementary Fig. S1). However, DNA-B and α-satellite molecules could not be amplified from the collected samples. These results implicated the presence of monopartite begomovirus complex. Consequently, the PCR amplified molecules were cloned in pJET1.2 cloning vector and the clones so obtained harbored the desired DNA product of ≈ 2.8 and ≈ 1.4 kb as confirmed by restriction digestion using EcoRV and KpnI for DNA-A and betasatellite, respectively (Supplementary Fig. S2 & S3A). Further, enzymatic digestion using restriction enzymes to screen the positive clones, revealed the presence of similar molecules in all the clones (Supplementary Fig. S3B & S4) and therefore, one representative clone from each sample was sent for sequencing. Since, the partial sequences of all the samples were similar only two DNA-A and one betasatellite molecule were fully sequenced and analyzed bioinformatically to study their phylogeny, divergence and recombination pattern.
Phylogenetic relatedness
The sequences so obtained were analyzed to find the homology with already submitted sequences in the database using NCBI BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Begomovirus sequences isolated from infected chilli (Bhut-Jolokia) plants (GenBank accession nos. MT886450 and MT886451) with leaf curl symptoms were found to be 99.6% similar with the ‘Indian’ strain of Cotton leaf curl Multan virus (CLCuMuV-IN; Accession no. MG373556) which was reported earlier from an infected hollyhock plant (family, Malvaceae). In addition to that, the betasatellite (GenBank accession no. MT886452) molecule cloned from infected chilli plant was found to be 99.8% identical with Tomato leaf curl Patna beta satellite (ToLCPaB, Accession no. EU862324).
Phylogenetic analysis by MEGA-X also signified the homology of isolated DNA-A (GenBank accession no. MT886450 and MT886451) with CLCuMuV-IN and betasatelite (GenBank accession no. MT886452) with ToLCPaB. The DNA-A sequences formed a separate clade with CLCuMuV-IN:ND:Hollyhock (GenBank accession nos. MG373556 and MG373551) and also found to share a common ancestor with CLCuMuV-PK:Cotton (GenBank accession no. EU365616) sequence (Fig. 2A). The whole cluster was more closely related to other cotton leaf curl virus species such as CLCuBuV (GenBank accession no. AM774303), CLCuKoV (GenBank accession no. GU385879), CLCuBaV (GenBank accession no. AY705380), and CLCuAlV (GenBank accession no. AJ002452), reported from Pakistan and India. However, CLCuMuV: Manipur isolates (MT886450 and MT886451) were distinctly related with ChiLCV isolates possibly because CLCuMuV was earlier known to infect members of Malvaceae family and may have emerged recently to broaden its host range and infect chilli belonging to Solanaceae. The associated betasatellite (GenBank accession no. MT886452) formed a clade with ToLCuPaB-IN:Bihar:tomato (GenBank accession no. EU862324) and also found to be closely related to ToLCuPaB-IN:Bihar:tobacco (GenBank accession no. HQ180393) (Fig. 3A). The entire cluster was also found to share a common origin with other betasatellites (GenBank accession no. JN663863, HM007112, JN663862 and EF190216) isolated from chilli. However, the newly isolated betasatellite (GenBank accession no. MT886452) was observed to be distinctly related to the betasatellites (GenBank accession nos. AY744380, JF502376, JF502377, AY083590, JF502393) isolated from cotton. Analysis of available viral sequence dataset also suggests the occurrence of CLCuMuV in several distinct hosts such as hollyhock, chilli, cotton, okra and potato, and amaranthus species [19].
Phylogenetic relatedness and color coded matrix of pairwise similarity scores of isolates of CLCuMuV-IN:Manipur:Chilli. A maximum-likelihood dendrogram generated through MEGA-X B Per cent nucleotide identity based heat map created using sequence demarcation tool (SDT), version 1.2. Tomato mottle virus (ToMoV) (GenBank accession no. AY965901) was chosen as an outgroup for the analysis
Phylogenetic relatedness and color coded matrix of pairwise similarity scores of cloned betasatellite, ToLCuPaB-IN:Manipur:Chilli. A Maximum-likelihood dendrogram generated through MEGA-X B Per cent nucleotide identity-based heat map created using sequence demarcation tool (SDT), version 1.2. Ageratum leaf curl disease associated betasatellite (GenBank accession no. FM164738) was considered as an outgroup for the analysis
The color coded matrix of pairwise similarity scores based on per cent nucleotide identity for DNA-A and betasatellite have been generated using SDT which further confirmed that the cloned CLCuMuV-IN:Manipur:Chilli (Accession nos. MT886450 and MT886451) belonged to the CLCuMuV (Fig. 2B) and the betasatellite (Accession no. MT886452) to ToLCuPaB (Fig. 3B).
GC plot analysis
Guanine-cytosine (GC) content refers to the proportion of guanine (G) and cytosine (C) in a given stretch of the genome. The innermost circle and bar represent the GC-plot graph with above-average (green color) and below average (purple color) GC content of the genome, with a window size of 100. In CLCuMuV genome (GenBank accession no. MT886450), low GC content was found to be in the AV1 region (Fig. 4A) whereas the GC distribution plot of ToLCuPaB revealed the presence of low GC content between the end of βC1 ORF and at the beginning of the A-rich region (Fig. 4B). Since, the low GC content region represents probable hot-spot for the recombination, both sequences were subjected to RDP to find the correlation between the GC content and potential recombinant breakpoints.
Genome analysis of isolated viral molecules. Outermost circle represents the nucleotide position in the circular genome, Inner colour code represents respective ORFs encoded by the DNA-A (A) and betasatellite (B), innermost circle and bar represent the GC-plot with above average (Green bar) and below average (Purple bar) GC content of the genome, with window size of 100 showing the highest and lowest possible regions of recombination respectively. This analysis was performed using Artemis DNA plotter version 18.1.0, (http://www.sanger.ac.uk/Software/Artemis). Illustrative representation of recombination patterns observed in the DNA-A (C) and betasatellite (D) genomes. Since the recombination events were similar in both the DNA-A sequences (MT886450 and MT886451), a single graphical image is shown. Details of probable recombination pattern is listed in Supplementary Table S3
Recombination analysis
The isolated DNA-A sequence represented possible recombination events in the AV1 region with high statistical significance. The analysis revealed that the isolated DNA-A molecules have CLCuMuV-PK (EU365616) and CLCuBuV-IN (KM070821) as major and minor parent, respectively. The p value ranges from 4.004 × 10−25 to 1.290 × 10−63 suggesting the significance of the predicted recombination events (Supplementary Table S3). The breakpoint positions were also found common between most of the sequences analysed signifying their common origin and relatedness. In the case of betasatellite, the recombination breakpoint was observed in the A-rich region. A pictorial representation of the data predicted by the RDP tool is shown (Fig. 4C, D). This deeper genome analysis by recombination and phylogenetic relatedness further suggests that the CLCuMuV isolates were present in neighbouring countries such as Pakistan and might have been introduced in India via the Western part which is now spreading to North-East region.
Evolutionary divergence analysis
Virus populations are very dynamic and influenced by several factors, such as population heterogeneity, geographical distributions, broad host range, and mixed infections. Estimation of the pairwise distance between the diverse begomovirus groups was carried out using ten distinct begomovirus isolates for comparison with the present CLCuMuV-IN:Manipur:Chilli (Accession no. MT886450). The data is signified by p value, with 95% confidence interval score (alpha = 0.05). Interestingly, minimum sequence diversity (17 nucleotides) was observed with CLCuMuV-PK:Cotton (Gen Bank accession no. EU365616) and maximum (74 nucleotide) with ChiLCV-PK IN:Chilli (GenBank accession no. JN663861) (Table 1). This further suggests the possibility of the cloned sequence isolated from infected plants of chilli var. Bhut-Jolokia to be more closely related to CLCuMuV.
Discussion
Bhut-Jolokia (a variety of hot chilli) is an important spice crop well-known globally for its highest pungency and great medicinal properties [3]. Bhut-Jolokia is an interspecific hybrid chilli pepper cultivated in few states of North-East India, including Assam, Nagaland, Manipur and Arunachal Pradesh. The pungency of Bhut-Jolokia is attributed to the presence of capsaicin and its analogs broadly known as capsaicinoids which possesses anti-inflammatory and antioxidant properties. These qualities resulted into its increased demand which is mostly hampered by leaf curl disease caused by begomoviruses. Recent studies have shown the effect of viral infection on secondary metabolite accumulation in plants [20]. In saffron which is highly appreciated for its colour and aroma, the presence of viral disease has been shown to modify the important secondary metabolite content responsible for its valuable properties [21, 22]. These reports suggest that virus infection not only affect the total yield loss but can also reduce the crop quality at the nutritional level in the surviving plants. Hence, a continuous monitoring of the infected field samples and thorough molecular analysis is the priority for developing management strategies and to prevent any epidemic.
The presence of CLCuMuV and its associated ToLCPaB satellite molecules from infected Bhut-Jolokia plants has been unveiled in the present study. The genome organization suggests the existence of old world monopartite begomoviral molecules at one of North-Eastern states of India, Manipur. Cloning, RFLP analysis and sequencing data further confirmed the presence of similar begomoviral molecules in all the samples collected at eight different locations in Manipur.
CLCuMuV is well-documented to cause cotton leaf curl disease (CLCuD) in several countries. In India, cotton plant infected with CLCuD was first reported at IARI, New Delhi in 1989. Subsequently, it was also isolated from other Malvaceous crops at North-Western states of India such as Rajasthan, Punjab and Haryana [23, 24]. Spreading of CLCuMuV from Pakistan to India and other adjoining countries are known earlier [25, 26]. It has been estimated that the cotton infecting begomoviruses are introduced in India from Africa, Pakistan, Philippines and China [27,28,29]. Furthermore, the genetic relatedness of the molecules using MEGA-X tool, taking into account the major chilli-infecting begomoviruses, suggested the homology of isolated CLCuMuV: Manipur isolates (GenBank accession nos. MT886450 and MT886451) with Cotton leaf curl Multan virus reported from New Delhi, India from an ornamental plant hollyhock [30] and the whole cluster was found more closely related to other cotton leaf curl virus isolates/strains such as CLCuBuV, CLCuKoV, CLCuBaV and CLCuAlV reported from Pakistan and India. As per the current ICTV recommendations [31] for the begomoviruses, the cloned components showed pairwise nucleotide identity above the threshold level (cutoff values of a novel species are ≥ 91%) and therefore, belong to another strain of CLCuMuV and ToLCPaB. However, CLCuMuV: Manipur isolates formed a separate clade from ChiLCV possibly because the former was known to infect the members of family Malvaceae, whereas chilli belongs to the Solanaceae family. This also highlights the spreading of reported CLCuMuV to a new host and an emergence of a novel and successful pathogen of chilli. Betasatellites are known to exhibit host-specific adaptation in crop plants and occasionally result in extending host range across families [32, 33].
In India, very few CLCuMuV (GenBank accession no. MF737345) isolates have been reported from infected chilli plants, however, a detailed analysis is still lacking. Evolution is often linked with recombination, and our data further suggests that CLCuMuV-PK might act as the major and CLCuBuV-PK as the minor parent donor for the recombination processes and the hot-spot region lies between the coat-protein (AV1) regions of the begomovirus genome. It has been already estimated and reported that the nucleotide substitution rate of CLCuMuV-CP (1.6 at codon position 1) was quite higher among other cotton leaf curl viruses, suggesting high mutation frequency in the CP region [27]. The most recent outbreak of CLCuD took place in 2011 [25], where Cotton leaf curl Kokhran virus (CLCuKuV) was found to be one of the putative parent donors in recombination analysis. Lately, an inter-species recombinant CLCuMuV-Rajasthan has been reported as the prime begomovirus to cause CLCuD in the North-West India [34]. These reports further suggest a prospective of intra or inter-strain species recombination that could be a potential threat for many economically important crops [35]. In case of the associated betasatellite molecule (ToLCuPaB), the recombination breakpoint was found in the A-rich region. Recombination event in Tomato leaf curl Patna betasatellite has previously been reported which further strengthens our observation [36]. Earlier reports suggest a viable mechanism of pseudorecombination in ToLCuPaB with ToLCGV DNA-A which resulted into successful establishment of infection in both host (tomato) and non-host (Nicotiana benthamiana) [36]. This proposes the possibility of similar event which may have resulted into infection even with a non-cognate CLCuMuV (DNA-A). A detailed insight into various betasatellites associated with chilli leaf curl disease causing begomoviruses [6] indicates a likelihood of ToLCuPaB helping the non-cognate CLCuMuV (DNA-A) to cause infection in the host. The GC content analysis also suggests the presence of below average GC content (the lowest) at the coat-protein region (AV1) in DNA-A molecule and the region spanning the A-rich region in betasatellite molecule, which further supports the possibility of viral genome to undergo recombination [37]. The low GC region has also been reported to be potential recombination site in human adenovirus which may facilitate the evolution [38]. The nucleotide sequence with high GC content depicts more stability because of the triple hydrogen bond and stacking interactions between the nitrogenous bases [39]. Higher the number of bonds, higher will be the energy required to break it. However, GC dinucleotide repeat is also linked with topology and orientation of DNA strand. In some viruses such as herpes simplex virus (HSV) genome, higher GC content is also found to be at the intergenic region [40] thus, suggesting a possible role in the virus evolution and its link with pathogenesis. Furthermore, the analysis of evolutionary divergence suggests that begomoviruses associated with leaf curl disease have significant genetic variations at their genome. These diverse begomovirus species not only provide enormous space for virus molecules to evolve but also allow a species to acclimatize a new host and a new environmental niche. Similar study has been demonstrated in SARS-CoV-2 genome sequences where nucleotide differences have been found between isolates reported form India and other countries [41]. This could be one of the factors for the spread of CLCuMuV in other crops. In recent years, CLCuMuV had been reported from many malvaceous (Okra, China rose), non-malvaceous (sunnhemp, jute), and solanaceaous (chilli/pepper) plants possibly due to broadening of its host-range. However, the role of viruliferous white-fly mediated virus transmission, anthropogenic activities, international trades of ornamental, and other crops plants cannot be ruled out. The spread of CLCuMuV among the economically important families of crop plants and its presence at distinct geographical areas is an alarming situation and needs further detailed investigation which may ultimately be used to develop successful and sustainable control strategies.
References
Bosland PW, Baral JB (2007) “Bhut Jolokia”—The world’s hottest known chile pepper is a putative naturally occurring interspecific hybrid. HortScience 42:222–224. https://doi.org/10.21273/hortsci.42.2.222
Purkayastha J, Alam SI, Gogoi H, Singh L (2012) Capsicum assamicum Sp. Nov. (Solanaceae), from Assam, Northeastern India. Ozean J Appl Sci 5:55–66
Roy A (2016) Bhut jolokia (Capsicum Chinense JAQC): a review. Int J Pharm Sci Res 7(3):882–889. https://doi.org/10.13040/IJPSR.0975-8232
Kumar S, Kumar S, Singh M et al (2006) Identification of host plant resistance to pepper leaf curl virus in chilli (Capsicum species). Sci Hortic 110:359–361. https://doi.org/10.1016/j.scienta.2006.07.030
Adams MJ, Antoniw JF (2006) DPVweb: a comprehensive database of plant and fungal virus genes and genomes. Nucleic Acids Res 34:D382–D385. https://doi.org/10.1093/nar/gkj023
Vinoth Kumar R, Singh AK, Singh AK et al (2015) Complexity of begomovirus and betasatellite populations associated with chilli leaf curl disease in India. J Gen Virol 96:3143–3158. https://doi.org/10.1099/jgv.0.000254
Singh AK, Kushwaha N, Chakraborty S (2016) Synergistic interaction among begomoviruses leads to the suppression of host defense-related gene expression and breakdown of resistance in chilli. Appl Microbiol Biotechnol 100:4035–4049. https://doi.org/10.1007/s00253-015-7279-5
Bhattacharyya D, Gnanasekaran P, Kumar RK et al (2015) A geminivirus betasatellite damages the structural and functional integrity of chloroplasts leading to symptom formation and inhibition of photosynthesis. J Exp Bot 66:5881–5895. https://doi.org/10.1093/jxb/erv299
Gnanasekaran P, Chakraborty S (2018) Biology of viral satellites and their role in pathogenesis. Curr Opin Virol 33:96–105. https://doi.org/10.1016/j.coviro.2018.08.002
Gnanasekaran P, KishoreKumar R, Bhattacharyya D et al (2019) Multifaceted role of geminivirus associated betasatellite in pathogenesis. Mol Plant Pathol 20:1019–1033. https://doi.org/10.1111/mpp.12800
Gnanasekaran P, Ponnusamy K, Chakraborty S (2019) A geminivirus betasatellite encoded βC1 protein interacts with PsbP and subverts PsbP-mediated antiviral defence in plants. Mol Plant Pathol 20:943–960. https://doi.org/10.1111/mpp.12804
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Rep 1:19–21. https://doi.org/10.1007/BF02712670
Chattopadhyay B, Singh AK, Yadav T et al (2008) Infectivity of the cloned components of a begomovirus: DNA beta complex causing chilli leaf curl disease in India. Arch Virol 153:533–539. https://doi.org/10.1007/s00705-007-0017-2
Birnboim HC, Doly J (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res 7:1513–1523
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Muhire BM, Varsani A, Martin DP (2014) SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE. https://doi.org/10.1371/journal.pone.0108277
Martin DP, Murrell B, Golden M et al (2015) RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:1–5. https://doi.org/10.1093/ve/vev003
Carver T, Thomson N, Bleasby A et al (2009) DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 25:119–120. https://doi.org/10.1093/bioinformatics/btn578
George B, Kumar RV, Chakraborty S (2014) Molecular characterization of chilli leaf curl virus and satellite molecules associated with leaf curl disease of amaranthus spp. Virus Genes 48:397–401. https://doi.org/10.1007/s11262-013-1027-7
Mishra J, Srivastava R, Trivedi PK, Verma PC (2020) Effect of virus infection on the secondary metabolite production and phytohormone biosynthesis in plants. 3 Biotech 10:547. https://doi.org/10.1007/s13205-020-02541-6
Parizad S, Dizadji A, Habibi MK et al (2019) The effects of geographical origin and virus infection on the saffron (Crocus sativus L.) quality. Food Chem 295:387–394. https://doi.org/10.1016/j.foodchem.2019.05.116
Moratalla-López N, Parizad S, Habibi MK et al (2021) Impact of two different dehydration methods on saffron quality, concerning the prevalence of saffron latent virus (SaLV) in Iran. Food Chem 337:127786. https://doi.org/10.1016/j.foodchem.2020.127786
Kumar A, Kumar J, Khan JA (2010) Sequence characterization of cotton leaf curl virus from Rajasthan: phylogenetic relationship with other members of geminiviruses and detection of recombination. Virus Genes 40:282–289. https://doi.org/10.1007/s11262-009-0439-x
Malathi VG, Renukadevi P, Chakraborty S et al (2017) Begomoviruses and their satellites occurring in India distribution, diversity and pathogenesis. In: Mandal B, Rao GP, Baranwal VK, Jain RK (eds) A century of plant virology in India. Springer, Singapore, pp 75–177
Rajagopalan PA, Naik A, Katturi P et al (2012) Dominance of resistance-breaking cotton leaf curl burewala virus (CLCuBuV) in Northwestern India. Arch Virol 157:855–868. https://doi.org/10.1007/s00705-012-1225-y
Datta S, Budhauliya R, Das B et al (2017) Rebound of cotton leaf curl multan virus and its exclusive detection in cotton leaf curl disease outbreak, Punjab (India) 2015. Sci Rep 7:1–14. https://doi.org/10.1038/s41598-017-17680-9
Saleem H, Nahid N, Shakir S et al (2016) Diversity, mutation and recombination analysis of cotton leaf curl geminiviruses. PLoS ONE 11:1–12. https://doi.org/10.1371/journal.pone.0151161
Sattar MN, Kvarnheden A, Saeed M, Briddon RW (2013) Cotton leaf curl disease—An emerging threat to cotton production worldwide. J Gen Virol 94:695–710. https://doi.org/10.1099/vir.0.049627-0
Sinha V, Kumar A, Bhatnagar D, Khan JA (2013) Association of cotton leaf curl multan virus and its satellite molecules with leaf curl disease of papaya in India. New Dis Rep 27:9. https://doi.org/10.5197/j.2044-0588.2013.027.009
Kumar M, Vinoth Kumar R, Chakraborty S (2020) Association of a begomovirus-satellite complex with yellow vein and leaf curl disease of hollyhock (Alcea rosea) in India. Arch Virol 165:2099–2103. https://doi.org/10.1007/s00705-020-04696-2
Zerbini FM, Briddon RW, Idris A et al (2017) ICTV virus taxonomy profile: geminiviridae. J Gen Virol 98:131–133. https://doi.org/10.1099/jgv.0.000738
Ranjan P, Singh AK, Kumar RV et al (2014) Host-specific adaptation of diverse betasatellites associated with distinct Indian tomato-infecting begomoviruses. Virus Genes 48:334–342. https://doi.org/10.1007/s11262-013-1031-y
Singh AK, Chattopadhyay B, Chakraborty S (2012) Biology and interactions of two distinct monopartite begomoviruses and betasatellites associated with radish leaf curl disease in India. Virol J 9:43. https://doi.org/10.1186/1743-422X-9-43
Biswas KK, Bhattacharyya UK, Palchoudhury S et al (2020) Dominance of recombinant cotton leaf curl multan-Rajasthan virus associated with cotton leaf curl disease outbreak in northwest India. PLoS ONE 15:1–19. https://doi.org/10.1371/journal.pone.0231886
Vinoth Kumar R, Singh D, Singh AK, Chakraborty S (2017) Molecular diversity, recombination and population structure of alphasatellites associated with begomovirus disease complexes. Infect Genet Evol 49:39–47. https://doi.org/10.1016/j.meegid.2017.01.001
Kumari P, Singh AK, Chattopadhyay B, Chakraborty S (2010) Molecular characterization of a new species of begomovirus and betasatellite causing leaf curl disease of tomato in India. Virus Res 152:19–29. https://doi.org/10.1016/j.virusres.2010.05.015
George B, Alam CM, Kumar RV et al (2015) Potential linkage between compound microsatellites and recombination in geminiviruses: evidence from comparative analysis. Virology 482:41–50. https://doi.org/10.1016/j.virol.2015.03.003
Robinson CM, Singh G, Lee JY et al (2013) Molecular evolution of human adenoviruses. Sci Rep 3:1812. https://doi.org/10.1038/srep01812
Ninh A (2013) Correlation between GC-content and palindromes in randomly generated sequences and viral genomes
Brown JC (2007) High G+C content of herpes simplex virus dna: proposed role in protection against retrotransposon insertion. Open Biochem J 1:33–42. https://doi.org/10.2174/1874091X00701010033
Devendran R, Kumar M, Chakraborty S (2020) Genome analysis of SARS-CoV-2 isolates occurring in India: present scenario. Indian J Public Health 64:S147–S155. https://doi.org/10.4103/ijph.IJPH_506_20
Acknowledgements
We gratefully acknowledge the financial support from the Department of Biotechnology, DBT (BT/PR16677/NER/95/246/2015).
Author information
Authors and Affiliations
Contributions
LS and KS performed the field inspection and collection of the infected samples. SC, SY and MK conceived the experiments and SY and MK performed the experiments and generated data. SC, SY and MK analyzed the data and drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human and animals rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yogindran, S., Kumar, M., Sahoo, L. et al. Occurrence of Cotton leaf curl Multan virus and associated betasatellites with leaf curl disease of Bhut-Jolokia chillies (Capsicum chinense Jacq.) in India. Mol Biol Rep 48, 2143–2152 (2021). https://doi.org/10.1007/s11033-021-06223-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06223-1