Abstract
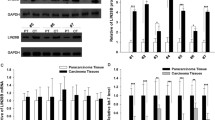
The let-7 family of microRNAs (miRNAs) are known to act as tumor suppressors and down-regulated in lung cancer. Recently, the RNA-binding protein Lin-28 was demonstrated to inhibit biogenesis of let-7 miRNAs by blocking both Drosha- and Dicer-mediated cleavage and accelerating decay of let-7 precursors. We selected NCI-H446 lung small cell lung cancer cell to determine whether it is broadly representative that Lin-28 can promote cell proliferation and affect cell cycle through negatively regulating let-7 biogenesis. Here, we showed that Lin-28 mRNA was up-regulated in NCI-H446 cell with a high c-Myc state. The result of real-time RT-PCR further indicated that pri-let-7a-1/7g and mature let-7g were remarkably down-regulated. The expression of lin-28 was down-regulated while the mature let-7g transcript was up-regulated inversely. The MTT assay indicated that the proliferation of lung cancer cells with lin-28 inhibition was signally impaired. The cells with lin-28 knockdown revealed a higher proportion of cells at G1/G0 phase and less at S phase. The results presented here demonstrate that induction of Lin-28 could mediate repression of let-7 family members, promote cell cycle progression and suppress cell proliferation.




Similar content being viewed by others
References
Yekta S, Shih IH, Bartel DP (2004) MicroRNA-directed cleavage of HOXB8 mRNA. Science 304:594–596
Vasudevan S, Tong Y, Steitz JA (2007) Switching from repression to activation: microRNAs can up-regulate translation. Science 318:1931–1934
Novotny GW, Sonne SB, Nielsen JE et al (2007) Translational repression of E2F1 mRNA in carcinoma in situ and normal testis correlates with expression of the miR-17-92 cluster. Cell Death Differ 14:879–882
Cullen BR (2004) Transcription and processing of human microRNA precursors. Mol Cell 16:861–865
Lu J, Getz G, Miska EA (2005) MicroRNA expression profiles classify human cancers. Nature 435:834–838
Du L, Pertsemlidis A (2010) MicroRNAs and lung cancer: tumors and 22-mers. Cancer Metastasis Rev 29:109–122
Landi MT, Zhao Y, Rotunno M et al (2010) MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res 16:430–441
Reinhart BJ, Slack FJ, Basson M et al (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403:901–906
Calin GA, Sevignani C, Dumitru CD et al (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 101:2999–3004
Inamura K, Togashi Y, Nomura K et al (2007) Let-7 microRNA expression is reduced in bronchioloalveolar carcinoma, a non-invasive carcinoma, and is not correlated with prognosis. Lung Cancer 58:392–396
Schultz J, Lorenz P, Gross G et al (2008) MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res 18:549–557
Shell S, Park SM, Radjabi AR et al (2007) Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA 104:11400–11405
Takamizawa J, Konishi H, Yanagisawa K et al (2004) Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 64:3753–3756
Kumar MS, Erkeland SJ, Pester RE et al (2008) Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA 105:3903–3908
Johnson SM, Grosshans H, Shingara J et al (2005) RAS is regulated by the let-7 microRNA family. Cell 120:637–647
Lee YS, Dutta A (2007) The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 21:1025–1030
Kumar MS, Lu J, Mercer KL et al (2007) Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 39:673–677
Johnson CD, Esquela-Kerscher A, Stefani G et al (2007) The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 67:7713–7722
Esquela-Kerscher A, Trang P, Wiggins JF et al (2008) The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle 7:759–764
Moss EG, Lee RC, Ambros V (1997) The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the Lin-4 RNA. Cell 88:637–646
Viswanathan SR, Daley GQ, Gregory RI (2008) Selective blockade of microRNA processing by Lin28. Science 320:97–100
Newman MA, Thomson JM, Hammond SM (2008) Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14:1539–1549
Piskounova E, Viswanathan SR, Janas M et al (2008) Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem 283:21310–21314
Heo I, Joo C, Kim YK et al (2009) TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138:696–708
Lehrbach NJ, Armisen J, Lightfoot HL et al (2009) LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol 16:1016–1020
Heo I, Joo C, Cho J, Ha M et al (2008) Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell 32:276–284
Rybak A, Fuchs H, Smirnova L et al (2008) A feedback loop comprising Lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol 10:987–993
Xu B, Zhang K, Huang Y (2009) Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. RNA 15:357–361
Chang TC, Zeitels LR, Hwang HW et al (2009) Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci USA 106:3384–3389
Viswanathan SR, Powers JT, Einhorn W et al (2009) Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet 41:843–848
Guo Y, Chen Y, Ito H et al (2006) Identification and characterization of Lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene 384:51–61
Balzer E, Moss EG (2007) Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol 4:16–25
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408
Molinari M, Mercurio C, Dominguez J et al (2000) Human Cdc25 A inactivation in response to S phase inhibition and its role in preventing premature mitosis. EMBO Rep 1:71–79
Moss EG, Tang L (2003) Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev Biol 258:432–442
Thomson JM, Parker J, Perou CM et al (2004) A custom microarray platform for analysis of microRNA gene expression. Nat Methods 1:47–53
Acknowledgments
This study was supported in part by research grants from the Natural Science Foundation of Ningbo (2009A610187), the Social Development Program of Ningbo (2007C10016), the Excellent Young Teachers Program of Education of Zhejiang Province, the K.C. Wong Magna Fund and Excellent Master Dissertation Cultivation Fund at Ningbo University.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pan, L., Gong, Z., Zhong, Z. et al. Lin-28 reactivation is required for let-7 repression and proliferation in human small cell lung cancer cells. Mol Cell Biochem 355, 257–263 (2011). https://doi.org/10.1007/s11010-011-0862-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-011-0862-x