Abstract
We tackle the problem of automatically filtering studies while preparing Systematic Reviews (SRs) which normally entails manually inspecting thousands of studies to identify the few to be included. The problem is modeled as an imbalanced data classification task where the cost of misclassifying the minority class is higher than the cost of misclassifying the majority class. This work introduces a novel method for representing systematic reviews based not only on lexical features, but also utilizing word clustering and citation features. This novel representation is shown to outperform previously used features in representing systematic reviews, regardless of the classifier. Our work utilizes a random forest classifier with the novel features to accurately predict included studies with high recall. The parameters of the random forest are automatically configured using heuristics methods thus allowing us to provide a product that is usable in real scenarios. Experiments on a dataset containing 15 systematic reviews that were prepared by health care professionals show that our approach can achieve high recall while helping the SR author save time.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Systematic Reviews (SRs) are literature surveys about a specific topic or treatment that seek to reach a conclusion in support of the hypothesis (i.e. treatment), or against it. What makes SRs unique is the strict guidelines and protocols under which SR authors need to operate and follow while researching and analyzing studies that are included in the review. In evidence based medicine (EBM), SRs are the pinnacle of evidence favoring a treatment or making the case against it based on a summary of the entire published work in that area.
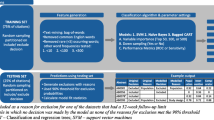
Preparing a SR is a very labor-intensive task. First, after identifying a research question for a given review, a comprehensive list of keywords is created and used to query several databases (e.g., PubMed and Web of Science) to find all the relevant studies. The next step is to review the title and abstract of each article and check it against a predefined set of inclusion criteria. Should the article pass the first filtering process, the full text of the article is retrieved and examined to judge whether it is to be included or excluded from the review. The labor-intensive nature of preparing SRs makes publishing reviews on a faster pace more challenging. For more information about systematic reviews, please refer to Cook et al. (1997).
While SRs are indispensable tools in EBM in particular, and medicine in general, they are susceptible to go out of date as soon as new key trials are published. Given that 75 trials are published per day (Bastian et al. 2010), the likelihood of a SR going out of date is ever increasing. A study for the survival rates of SRs (Shojania et al. 2007) found that the median number of years before a SR goes out of date is 5.5 years. However, 23 % of SRs needed update within 2 years of publishing (Shojania et al. 2007). The need for constant update, along with the need for new SRs in areas that are not covered yet makes producing SRs and keeping them up to date in a timely fashion much harder. Therefore, reducing the workload on SR authors is imperative to shorten the timespan of producing new reviews.
One key area at which SR authors might use some help is the screening of articles for inclusion in the review. There are two steps for screening that are conducted sequentially, the abstract stage and the full text stage. The number of articles reviewed in each SR varies significantly, ranging from hundreds to thousands and even tens of thousands. Table 1 shows the number of articles screened for 15 SRs conducted by the Oregon Evidence Based Practice Center (EPC), Southern California EPC, and Research Triangle Institute/University of North Carolina (RTI/UNC) EPC.Footnote 1 The percentage of included articles at the full text level (hereafter inclusion means included at the full text level) ranges from 0.5 to 21.7 %, however it is below 15 % for 13 out of the 15 SRs. This highly skewed distribution presents a challenge for machine learning approaches to identify the correctly included articles. Yet, it is desirable to reduce the number of excluded articles (i.e. irrelevant) that the reviewer has to go through before identifying all the relevant articles. In addition, missing an included article is very costly as it may sway the conclusion of the SR.
In this work we describe one part of Rayyan, a system for health professionals for preparing systematic reviews. More specifically, we describe a new label prediction module that is under development within Rayyan which provides relevance prediction for the users once they label enough studies. Previous work on SR inclusion prediction has mostly used lexical and syntactic features to represent clinical trials in a bag-of-words model. We extend this representation by introducing a novel set of features that incorporate citation information for each clinical trials. The intuition here is that co-cited articles are similar to a certain degree, therefore if one article is included then the other co-cited articles have a good chance of being included as well. We also use word clustering as computed by Brown clustering (Brown et al. 1992) algorithm to find clusters representing collections of similar words. As we will show later, this combination of novel features outperforms conventional textual features regardless of the classification algorithm.
In this paper, we use random forests (Breiman 2001), an ensemble classification method, to identify the included articles from the excluded ones. We devise methods and heuristics to tackle the imbalanced data problem, and induce parameters a priori for the random forest. The introduced heuristics for tuning classifier parameters are essential in creating a useful system for the users. It is also an improvement over existing work which has always reported best performance with different parameters for each dataset without having a unified way of configuring the parameters. This limitation presents challenges when deploying real systems. We are unaware of any previous work that utilized the citation information or word clustering in classifying clinical trials for inclusion in SRs. The main contribution of this work can be summarized in the following: (1) We introduce a novel set of features that are shown to be good predictors of SR inclusion decisions. (2) We design a classification framework based on minimizing the expected loss of a Random Forest classifier wherein all parameters of the models are chosen based on formulas. (3) We demonstrate how this model can be used to achieve near 100 % recall in identifying relevant studies when updating an already published systematic review. (4) The proposed methods are to be added to the Rayyan system which is used for preparing systematic reviews by health care professionals.
The remainder of this paper is organized as follows. Section 2 surveys the related work, and Sect. 3 describes the used dataset briefly. In Sect. 4 we describe the features used in representing clinical trials. Our approach is introduced in Sect. 5. Section 6 reports the experiments and results. We finally conclude in Sect. 7 and discuss future work.
2 Related work
There has been a large body of work about applying machine learning methods to aid creating and updating SRs, especially classifying articles into include and exclude classes. Cohen et al. (2006) were the first to investigate reducing the workload of SR authors using machine learning methods. They contributed a dataset comprising 15 SRs along with the inclusion and exclusion judgments at the abstract and article level. Their approach used a voting perceptron algorithm with varying learning weights to penalize for misclassifying the include class (hereafter, include class will be used interchangeably with positive class and minority class). They were able to achieve 95 % recall on the include class, with different weights for each SR. There is, however, no universal way for choosing the best weight across all reviews beforehand. Another contribution of their work is the measure by which the classification performance was evaluated. They introduced the Work Saved Over random Sampling (WSS) which is the percentage of articles that the reviewer does not have to review as a result of automatic classification, compared to random sampling.
where TN is the number of true negatives, FN is the number of false negatives, and N is the total number of instances in the dataset. Recall refers to the recall of the positive class. For example, if the total number of studies considered for a given SR \(N=100\), out of which 10 are to be included, and the classifier identified 60 true negative studies (TN) and 1 false negative studies (FN), whereas (\(TP=9\)) and \(FP=30\), then WSS would be 51 % (the recall is 0.9). So just by random sampling one would expect saving of 10 % at the level of 90 % recall. Note that this can be achieved by keeping 90 % of the data only, hence saving 10 % of the work while expecting a recall of 90 %. The WSS measure simply adjusts the savings by subtracting the expected saving achieved by mere sampling.
Later, Cohen (2008) studied the performance of Support Vector Machine (SVM) classifier using different collection of features both textual and conceptual. Their best performance was obtained using a combination of unigram, bigram terms of titles and abstracts, along with MeSH terms. In that work, they reported the performance using Area Under the Curve (AUC) instead of precision, recall, and WSS.
Matwin et al. (2010) used Complement Naive Bayes, a variation of the traditional Naive Bayes classifier, to predict the inclusion and exclusion decisions using textual features from the abstracts of studies. Their approach assigns a SR dependent weight multiplier for the features in order to obtain competitive accuracy. The weight multiplier needs to be explored a priori, but there is no rule to assign the weight, instead multiple values are tested and the one obtaining the best performance is reported in the work. They tested their approach on the same data set as in Cohen et al. (2006).
A different approach utilizing active learning was developed by Wallace et al. (2010a, b, 2011). In the active learning approach, instead of randomly splitting the data into training and testing set, the system chooses the instances on which it is mostly confused, and asks the human to label them. Hence, the more informative examples are labeled by the human which leads to less time being spent on labeling while sustaining the required accuracy. Miwa et al. (2014) have also explored different strategies of active learning when combined with Latent Dirichlet Allocation (LDA) to automatically screen studies for inclusion. While they found active learning to be effective for complex topics, the efficiency was rather limited. Others have explored linked data and semantic based approaches to learn included and excluded studies (Tomassetti et al. 2011; Jonnalagadda and Petitti 2013; Kouznetsov et al. 2009).
Recently, Cohen et al. (2012) have considered the problem of updating a SR. Their work predicts the clinical trials which should be included in an update of an existing SR. While predicting the inclusion decisions for the updates is no different than when preparing the SR for the first time from the classifier stand point, it however introduces the problem of concept drifting where the key information and interest of the preparer drift over time.
3 Dataset
The dataset used in this work was created by Cohen et al. (2006). It contains inclusions and exclusion annotations for 15 systematic reviews that were prepared by health care professionals from multiple centers. Table 1 shows the number of included and excluded studies at both the abstract and full text triage stages for each of the systematic reviews. For each systematic reviews, the list of examined articles were recorded in Endnote files. The authors of Cohen et al. (2006) processed all the end note files and matched the examined article’s metadata with Medline database by associating each article with a Pubmed Id. Then, for each systematic review, the list of included and excluded Pubmed Ids are provided. This is one of the few datasets that contain real systematic review triaging annotations. It was later used in multiple studies (Matwin et al. 2010; Cohen 2008).
4 Features and representation
The features that we investigated in classifying included/excluded articles can be classified into three classes. Textual features, citations features, and word co-occurrence information (Brown Clustering). Each distinct value of a given feature is a dimension in the vector space, such that each term, citation, or Brown cluster corresponds to a dimension. The weights of each feature are kept as binary. For example, a given study X is represented as a vector where each dimension corresponds to a feature from one of the classes listed below \(X = (x_{t_1},\ldots ,x_{t_k},x_{c_1},\ldots ,x_{c_j},x_{b_1},\ldots ,x_{b_m})\), where \(x_{t_i}\) refers to a textual feature, \(x_{c_l}\) refers to a citation feature, and \(x_{b_r}\) refers to Brown clustering feature.
4.1 Textual features
The title and abstract of each article is used to generate word level n-gram features. We considered unigram and bigram only as previous research has shown that unigram and bigram features outperform higher-order n-grams (Cohen 2008). Along with the title and abstract, we use MeSH terms and the publication type of the article as classified by Pubmed. Each possible MeSH term and publication type corresponds to one textual feature \(x_t\). We make a distinction between the same words appearing in the title and the abstract such that an occurrence at the title level corresponds to a different feature than occurrence at the abstract level. Apache Lucene Standard AnalyzerFootnote 2 is used to tokenize and generate n-grams while removing stop words.
4.2 Co-citations
As the building blocks of any SR comprises a collection of published trials and articles, this published work receives citations from other studies. The citation data constitute valuable information about each article which cannot be captured by the textual content. Citations are used to construct citation networks that are used to find related studies based on co-citation behavior (Small 1973). We define co-citation as follows: Articles A and B are co-cited if there exist an article C such that C cites both A and B. To the best of our knowledge, this is the first work to include citation information to represent articles for the purpose of SR screening. The intuition here is that co-cited articles have a degree of similarity for them to be co-cited, therefore it would be more likely to include a given article if it is co-cited with an already included article. We obtain the list of incoming citations for every article that is considered for inclusion in Cohen et al. (2006) dataset using Google Scholar. For each article, we submit a query using its DOI and the list of incoming citations, if any, is stored. In case the search using DOI fails, we use the article’s title. For all the articles found in the dataset (Cohen et al. 2006), we obtain the incoming citations using Google Scholar. Overall, we collect 628 thousands citations for 59 % of the papers, where the remaining 41 % either did not have citations in Google Scholar, or were not found on the first search page. In the future we plan on exploring other sources of citations such as Web of Science.
4.3 Brown clustering
Often in many NLP applications, data sparsity creates a problem as features from the testing data might not have appeared in the training set. Features extracted using word clustering aims to tackle this problem by representing each word as a code that refers to a cluster of related words that appear in a large corpus. When a new word appears in the test set, the cluster which it belongs to might have appeared in the training set before.
We employ the Brown clustering algorithm (Brown et al. 1992) to infer clusters containing words that are related to each other. Brown’s algorithm applies hierarchical clustering on the bigrams of a corpus, generating a binary tree of clusters where each word is assigned to a cluster. The tree is encoded using Huffman code resulting in each cluster being represented as a string of zeros and ones, and hence each word belongs to a cluster whose representation is a binary code.
To create Brown clustering codes for a relevant corpus of the problem we are working on, we obtained more than 300,000 abstracts of clinical trials from PubMed. The sentences of the abstracts were split and tokenized using Stanford NLP parser, resulting in 2.5 million sentences accounting for 1.3 million unique words. We run the Brown clustering algorithm on the 2.5 million sentences to generate 1000 clusters.
5 Approach
We use Random Forests (RF) (Breiman 2001) as a classification method to predict the included studies from the excluded ones. RF is an ensemble method which grows multiple unpruned decision trees, each of which is grown on a bootstrap sample of the training data where only a random subset of the features is used to split each node in the decision tree. At prediction time, the class receiving the majority of the votes from the individual trees is the predicted class of the forest. The bootstrap nature and random feature selection of the random forest give it nice theoretical and practical properties that make it perform well on a variety of classification problems (Caruana and Niculescu-Mizil 2006). It also compares favorably with approaches like AdaBoost (Freund et al. 1996) for generalization of error rate.
Random Forests, just like most classifiers, were not designed to deal with imbalanced data (Chen et al. 2004). However, screening citations for inclusion in SRs is a typical example of imbalanced data as the number of included articles is much smaller than the number excluded articles. In addition, missing an included article is very costly as it may sway the conclusion of the SR. Traditionally, there have been two methods to deal with imbalanced data in classification problems. The first approach is cost sensitive classification where the cost of mispredicting the minority class is higher than the cost of mispredicting the majority class. In this approach, the classifier will output the class which has the lowest expected misprediction cost. The second approach relies on over-sampling the minority class or under-sampling the majority class to create a balanced dataset from which the classifier learns.
In this work, we use a cost sensitive classification approach that is inspired by the MetaCost algorithm (Domingos 1999), where we assign asymmetric weights to misclassifying each class. Let Cost(m|n) be the cost of predicting class m when the true class is n, where \(m, n \in \{I, E\}\) and I, E denote include and exclude respectively. Therefore, Cost(I|E) is the cost of predicting include when the true label is exclude, and Cost(E|I) is the cost of predicting exclude when the true label is include.
By definition the \(Cost(m|m) =0\) where \(m \in \{I, E \}\). Also let P(I) be the probability of including the article, and P(E) be the probability of excluding the article where \(P(I)+P(E)=1\). The probabilities P(I) and P(E) are estimated as the fraction of trees in the random forest voting in favor of include and exclude, respectively. Given the previous variables, we compute the expected loss of predicting each class as (the conditional risk Duda and Hart (1973)):
and similarly:
Then, the predicted class is given by the Bayes optimal prediction :
The Bayes optimal prediction is guaranteed to achieve the lowest possible overall cost (Domingos 1999).
The values of the misclassification cost for the include class is assigned using the following heuristic:
where c is a constant that is set to 2 empirically, and the ratio r is computed from the training data. This approach for setting costs has been reported previously (Domingos 1999), and our experiments with multiple values for c arrived at a value of 2. Note that all empirical settings were based on the training data. Assuming the dataset is shuffled uniformly before splitting into training and testing datasets, the ratio r is expected to be the same in the two parts. The value of Cost(I|E) is left as 1, which is the default value when a non cost sensitive classifier is used.
Random Forest requires configuring two crucial parameters, the number of trees to grow, and the number of random features to consider at each node. We seek to devise heuristics based on formulas to assign values to these parameters such that these heuristics are applicable to any SR. Hence, the formula has to depend on the dataset only. We set the number of trees to be:
where k is a constant that is set to 0.3 empirically, and n is the number of articles considered for inclusion in a given SR (the number of data points in training and testing). The upper bound 1000 is used to limit the number of trees in case the number of data points is too large, thus keeping the resources of the system in control. As for the number of features to split upon, it is assigned as follows:
where M is the total number of features in the dataset. This heuristic, which is the default configuration of WEKA (Hall et al. 2009), was found to obtain error rate lower than \(\sqrt{M}\) that is suggested by Breiman (2002).
6 Experiments and discussions
We have conducted experiments to study the accuracy of the classification approach using different subsets of the features. Along with that, we studied the performance at different splits of training and testing datasets. For a system that is to be deployed for real use, we need to find out what is the minimum required size of a training dataset to produce satisfactory predictions. In the third experiment, we focus on the SR update process where the decisions on an already published review can be used to predict what should be included in a future update.
6.1 Classification
In the first experiment, we study the performance of our random-forest-based classifier using the different classes of features that we have introduced earlier. Previous research (Cohen 2008) showed that unigrams and bigrams are optimal features for a SVM classifier. However, as we introduce new features (e.g., citations), bigrams may not be the best predictor.
For each dataset, we perform a \(5\times 2\) cross validation to be consistent with the related work we compare against.Footnote 3 In \(5 \times 2\) cross validation, the dataset is first split in half with one half used for training and the second used for testing. In these experiments, split is carried out with stratification, similar to previous work. Then the roles of each half are switched. This process is repeated 5 times, resulting in 10 estimates that are averaged at the end.
In Table 2, we report the obtained recall along with the corresponding WSS for each SR. There are several experiments, each conducted with different combinations of features. The different features are title and abstract unigrams (Uni), title and abstract bigrams (Bi), citation information (Cite), Huffman codes from Brown clustering (BC), and length 12 and 16 prefixes of Huffman codes, along with the full code, generated from Brown clustering (SubBC). While we have experimented with all possible combinations, we report mostly on the unigram-based features as they were the most competitive in terms of recall. All of the models included publication type and MeSH term features by default. From Table 2, we find that the combination of unigrams and Brown clustering codes (Uni+BC) achieved the highest recall in 7 out of the 15 systematic reviews, tied with unigram and Brown clustering code prefixes (Uni+SubBC). However, the unigram and citation (Uni+Cite) model had recall values very close to the (Uni+BC) model. In some cases the difference was less than 0.001. We perform statistical significance test on the combination of features where we found that these models are statistically insignificant, hence we adopt the (Uni+Cite) model for the rest of the paper.
We compare the obtained WSS against values of WSS reported by other approaches in Table 3. The included approaches are voting perceptron (VP) (Cohen et al. 2006), Complement Naive Bayes (CNB) (Matwin et al. 2010), and the citations + unigram based random forest (RF). The recall column reports the recall value for the positive class obtained by RF. For the VP approach, the authors report WSS at different values of recall. Therefore, comparing with VP is carried by picking the value of WSS corresponding to the closest recall obtained by RF. Note that WSS values for CNB are for recall of 95 % and not for the same level of recall obtained by RF. Since our approach does not depend on tweaking variables to obtain different values of recall, we cannot always guarantee a recall of 95 %, as sometimes it can be higher (8 out of 15 in the datasets had recall \(>\) 95 %), or slightly smaller (4 out of 15). Generally, higher values of recall are correlated with lower values of WSS. Overall, not only is our approach capable of obtaining recall higher than 95 %, but also outperforms CNB in 5 out of the 15 datasets, while being comparable in 3 others. Note that the CNB values are selected for the weights yielding the highest recall, where these weights cannot be chosen before hand. When compared against VP, our RF based classifier is outperforming it in 13 out of the 15 datasets.
To compare with the SVM approach reported in Cohen (2008) we compute AUC for our RF model because Cohen (2008) only reported AUC. We compare RF classifier with different feature sets against the reported SVM approach that uses title and abstract n-grams along with mesh terms. We report the AUC values for our (Uni+Cite) random forest model in Table 3. When statistical significance test is performed to compare our AUC values against the ones reported in Cohen (2008), our RF model ranked in the first rank group, while the model based on Cohen (2008) was in the third rank group when all possible combinations of features were tested. Therefore, a model based on our features will have higher AUC than Cohen (2008). We also notice that a SVM model with citations and/or Brown clustering features yields statistically insignificant results when compared to a RF model on all the datasets, despite having slightly lower AUC value. This suggests that the contribution is resulting from the features more than the classifier when optimizing for AUC. Therefore, we compare the contribution of each feature groups to the accuracy of the classifier. Table 4 lists the AUC of each possible combination of the feature groups after grouping statistically insignificant results together using Wilcoxon tests. All combinations were tried to get the best feature set. The number of non-zero combinations equals to \(2^6 - 1 =63\). Antihistamines and SkeletalMuscleRelaxants reviews were excluded as they are considered outliers because of having small absolute number of included studies, as pointed out by Cohen (2011) and Matwin et al. (2010).
6.2 Required training size
In the previous experiments, we have examined the recall and WSS of the classifier based on a \(5 \times 2\) fold cross validation where 50 % of the data being used for training. However, we would like to find the smallest percentage of training data with which the classifier is making accurate predictions. We vary the size of the training dataset to be 10, 20, 30, and 40 % of the total dataset size. For each training size percentage, five splits are created and the final result corresponds to the average of these 5 splits. While the approach we devised relies on having the ratio of include to exclude articles reasonably similar in the training and testing dataset, in this experiment we relax this constraint and study the performance when the training and testing datasets are split without stratification. This scenario arises in real life when SR preparers might review articles that are sorted by some measure of similarity, or the case when a reviewer starts reviewing articles before compiling the entire list of candidate articles. Therefore, in this experiment we compare the performance of the classifier at various percentages of splits for both a stratified sample and an non-stratified sample. For each sampling approach, five splits are carried at each level and the average performance over these five splits is reported. Note that in case of an non-stratified split, the training data may have zero positive examples, hence calculating the ratio r that computes the misclassification penalty is not possible. When a split results in zero positive examples in the training data, the split is ignored—one such split was encountered per dataset at most.
In Figs. 1 and 2, values of recall and WSS for stratified and random splitting are plotted against different percentages of training data size, where each sub plot represents a specific SR from our sample. S and R denote stratified and non-stratified sampling, respectively. In 12 out of the 15 SRs, the recall and WSS values start to converge at training data set split of 30 %. While recall tends to converge at lower percentages of training, increasing the training size to 30 % is more beneficial to WSS than recall. That is, having more training data is reducing the false positive rate, hence saving the time of SR authors. Interestingly, previous research that applied active learning to select which articles to tag reported needing up to 30–40 % of the entire dataset to be able to achieve recall as high as 1 (Wallace et al. 2010b) (Albeit working with a different dataset that contains three SRs only, their results about the required percentage of training data are informative here). In the Web application http://rayyan.qcri.org/ that we deployed, the application simply decides when it is confident enough to show predictions by running a cross validation on the already tagged articles.
6.3 Updating systematic reviews
As SRs go out of date, it is crucial to find and include the newly published studies that might change the recommendation of the review. SR authors are now presented with a collection of studies that were published after the review went to print so they would sift through the titles and abstracts to identify potential relevant studies. A classification algorithm can be used here to filter the list of newly published studies based on the inclusion and exclusion decisions made while creating the review in the first place. This, of course, depends on storing the decisions for the studies used in the initial review. We, however, were unable to obtain a dataset that records the list of studies used when creating a SR, along with the list of studies considered in the update phase. Automatic classification techniques are more acceptable here because the reviewers are not asked to manually tag studies. Instead, the tags generated while creating the original review are used to train the model.
To model this, we set up the following experiment. For each of the 15 SRs described earlier, we mimic an update scenario by assuming that all articles published on or before 2001 were used for creating the original review, and the articles published between 2002 and 2004 are used for a hypothetical update taking place in 2005. The dates were chosen such that there is enough body of work published after the original review goes online that mandates an update. Furthermore, three years period is a reasonable time span after which SR might need an update based on Shojania et al. (2007). The number of included and excluded studies published before 2002, and after it is reported in Table 5. Note that the dataset used here contains studies from 1991 to 2004 only.
We use the random forest classifier with unigram and citation features. The results are presented in Table 5. In 11 out of the 15 SRs, we are able to achieve a recall of 1 with WSS values ranging from 0.09 up to 0.52. By examining the datasets closely, we find that in SRs 2 and 14 the number of included studies published after 2001 is equal to or larger than the number of included articles published before 2002 indicating that the timeline split is a good fit for these reviews. This can explain the lower than 1 recall incurred for these two reviews. Overall, the average WSS was 0.3671, meaning that with a recall of nearly 1, the classifier is saving 36 % of the SR preparer’s time. This can translate into hours, if not days, worth of work.
6.4 Integration with Rayyan
Rayyan (http://rayyan.qcri.org/) is a web-enabled application that helps systematic review authors expedite their work. Authors upload a list of studies obtained from searches on different databases and start screening them for inclusion and exclusion. Using different facets, e.g., extracted MeSH terms, keywords for inclusion, keywords for exclusion, journals, authors, and year of publication, they navigate through their citations and filter them to focus on those they want to exclude or exclude. They can also browse a similarity graph for the studies based on different attributes such as titles and authors. As they browse and filter on studies, they select those to include or exclude. They can also label them for easy reference or for reporting the reason for inclusion or exclusion. Once they have excluded and included enough studies, the prediction module is run and suggestions on undecided studies are returned to the users who will then make the actual decisions. When updating a review, users simply upload a new set of studies and Rayyan will then provide suggestions on these. Other features of Rayyan include the ability to have multiple collaborators work on the same review, uploading multiple files (list of studies) to the same review, and copying of studies across reviews.
7 Conclusion and future work
In this work we have introduced an ensemble-based approach using Random Forests to identify relevant studies for SRs. We model the problem as an imbalanced classification task, and assign asymmetric weights for misclassifying each class then minimize the expected loss. Our proposed approach utilizes a unique collection of features that uses outside information including co-citations and word embeddings to predict the relevant studies. We devise methods and heuristics to assign costs for misclassifying each class using the training data, thus allowing the parameters of the classifier to be set automatically.
Experiments on a dataset containing 15 SRs shows that we are able to identify all the relevant studies with recall in the upper 90s, while saving the SR preparer’s time by 31 % on average. We have also simulated the validity of our approach on updating SRs, and in 12 out of 15 reviews we were able to obtain a recall of 1.
In the future we will be exploring other document features that will potentially increase the recall, especially other methods for computing word embeddings.
Notes
References
Bastian, H., Glasziou, P., & Chalmers, I. (2010). Seventy-five trials and eleven systematic reviews a day: How will we ever keep up? PLoS Medicine, 7(9), e1000,326.
Breiman, L. (2001). Random forests. Machine Learning, 45(1), 5–32.
Breiman, L. (2002). Manual on setting up, using, and understanding random forests v3. 1. http://oz.berkeley.edu/users/breiman/Using_random_forests_V3.1.
Brown, P. F., Desouza, P. V., Mercer, R. L., Pietra, V. J. D., & Lai, J. C. (1992). Class-based n-gram models of natural language. Computational Linguistics, 18(4), 467–479.
Caruana, R., & Niculescu-Mizil, A. (2006). An empirical comparison of supervised learning algorithms. In Proceedings of the 23rd international conference on machine learning, pp. 161–168. ACM, New York.
Chen, C., Liaw, A., & Breiman, L. (2004). Using random forest to learn imbalanced data. Berkeley: University of California.
Cohen, A. M. (2008). Optimizing feature representation for automated systematic review work prioritization. In AMIA annual symposium proceedings, vol. 2008, p. 121. American Medical Informatics Association, New York
Cohen, A. M. (2011). Performance of support-vector-machine-based classification on 15 systematic review topics evaluated with the wss@ 95 measure. Journal of the American Medical Informatics Association, 18(1), 104–104.
Cohen, A. M., Ambert, K., & McDonagh, M. (2012). Studying the potential impact of automated document classification on scheduling a systematic review update. BMC Medical Informatics and Decision Making, 12(1), 33.
Cohen, A. M., Hersh, W. R., Peterson, K., & Yen, P. Y. (2006). Reducing workload in systematic review preparation using automated citation classification. Journal of the American Medical Informatics Association, 13(2), 206–219.
Cook, D. J., Mulrow, C. D., & Haynes, R. B. (1997). Systematic reviews: Synthesis of best evidence for clinical decisions. Annals of Internal Medicine, 126(5), 376–380.
Domingos, P. (1999). Metacost: A general method for making classifiers cost-sensitive. In Proceedings of the fifth ACM SIGKDD international conference on Knowledge discovery and data mining, pp. 155–164. ACM, New York
Duda, R. O., Hart, P. E., et al. (1973). Pattern classification and scene analysis (Vol. 3). New York: Wiley.
Freund, Y., Schapire, R.E., et al. (1996). Experiments with a new boosting algorithm. In ICML, vol. 96, pp. 148–156
Hall, M., Frank, E., Holmes, G., Pfahringer, B., Reutemann, P., & Witten, I. H. (2009). The weka data mining software: An update. ACM SIGKDD Explorations Newsletter, 11(1), 10–18.
Jonnalagadda, S., & Petitti, D. (2013). A new iterative method to reduce workload in systematic review process. International Journal of Computational Biology and Drug Design, 6(1), 5–17.
Kouznetsov, A., Matwin, S., Inkpen, D., Razavi, A. H., Frunza, O., Sehatkar, M., Seaward, L., & OBlenis, P. (2009). Classifying biomedical abstracts using committees of classifiers and collective ranking techniques. In Advances in artificial intelligence, pp. 224–228. Berlin: Springer
Matwin, S., Kouznetsov, A., Inkpen, D., Frunza, O., & O’Blenis, P. (2010). A new algorithm for reducing the workload of experts in performing systematic reviews. Journal of the American Medical Informatics Association, 17(4), 446–453.
Miwa, M., Thomas, J., OMara-Eves, A., & Ananiadou, S. (2014). Reducing systematic review workload through. Journal of Biomedical Informatics, 51, 242–253.
Raeder, T., Hoens, T. R., & Chawla, N. V. (2010). Consequences of variability in classifier performance estimates. In IEEE 10th international conference on data mining (ICDM), 2010, pp. 421–430. IEEE.
Shojania, K. G., Sampson, M., Ansari, M. T., Ji, J., Doucette, S., & Moher, D. (2007). How quickly do systematic reviews go out of date? A survival analysis. Annals of Internal Medicine, 147(4), 224–233.
Small, H. (1973). Co-citation in the scientific literature: A new measure of the relationship between two documents. Journal of the American Society for information Science, 24(4), 265–269.
Tomassetti, F., Rizzo, G., Vetro, A., Ardito, L., Torchiano, M., & Morisio, M. (2011). Linked data approach for selection process automation in systematic reviews. In 15th Annual Conference on Evaluation & Assessment in Software Engineering (EASE 2011), pp. 31–35. IET.
Wallace, B. C., Small, K., Brodley, C. E., & Trikalinos, T. A. (2010). Active learning for biomedical citation screening. In Proceedings of the 16th ACM SIGKDD international conference on Knowledge discovery and data mining, pp. 173–182. ACM, New York
Wallace, B. C., Small, K., Brodley, C. E., & Trikalinos, T. A. (2011). Who should label what? Instance allocation in multiple expert active learning. In SDM, pp. 176–187.
Wallace, B. C., Trikalinos, T. A., Lau, J., Brodley, C., & Schmid, C. H. (2010). Semi-automated screening of biomedical citations for systematic reviews. BMC Bioinformatics, 11(1), 55.
Author information
Authors and Affiliations
Corresponding author
Additional information
Work done while all authors were at QCRI.
Editors: Byron Wallace and Jenna Wiens.
Rights and permissions
About this article
Cite this article
Khabsa, M., Elmagarmid, A., Ilyas, I. et al. Learning to identify relevant studies for systematic reviews using random forest and external information. Mach Learn 102, 465–482 (2016). https://doi.org/10.1007/s10994-015-5535-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10994-015-5535-7