Abstract
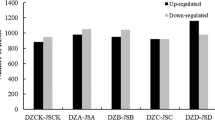
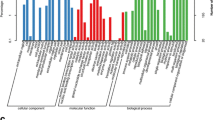
Medicago sativa L. (alfalfa) ‘Zhaodong’ is an important forage legume that can safely survive in northern China where winter temperatures reach as low as −30 °C. Survival of alfalfa following freezing stress depends on the amount and revival ability of crown buds. In order to investigate the molecular mechanisms of frost tolerance in alfalfa, we used transcriptome sequencing technology and bioinformatics strategies to analyze crown buds of field-grown alfalfa during winter. We statistically identified a total of 5605 differentially expressed genes (DEGs) involved in freezing stress including 1900 upregulated and 3705 downregulated DEGs. We validated 36 candidate DEGs using qPCR to confirm the accuracy of the RNA-seq data. Unlike other recent studies, this study employed alfalfa plants grown in the natural environment. Our results indicate that not only the CBF orthologs but also membrane proteins, hormone signal transduction pathways, and ubiquitin-mediated proteolysis pathways indicate the presence of a special freezing adaptation mechanism in alfalfa. The antioxidant defense system may rapidly confer freezing tolerance to alfalfa. Importantly, biosynthesis of secondary metabolites and phenylalanine metabolism, which is of potential importance in coordinating freezing tolerance with growth and development, were downregulated in subzero temperatures. The adaptive mechanism for frost tolerance is a complex multigenic process that is not well understood. This systematic analysis provided an in-depth view of stress tolerance mechanisms in alfalfa.









Similar content being viewed by others
Abbreviations
- Nt:
-
NCBI nonredundant nucleotide
- Nr:
-
NCBI nonredundant protein
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- KOG:
-
EuKaryotic Orthologous Groups
- DEGs:
-
Differentially expressed genes
- CBF:
-
C-repeat binding factor
- qPCR:
-
Quantitative real-time polymerase chain reaction
- TF:
-
Transcription factor
References
Achard P, Gong F, Cheminant S, Alioua M, Hedden P, Genschik P (2008) The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20:2117–2129. doi:10.1105/tpc.108.058941
Baek KH, Skinner DZ (2003) Alteration of antioxidant enzyme gene expression during cold acclimation of near-isogenic wheat lines. Plant Sci 165:1221–1227. doi:10.1016/S0168-9452(03)00329-7
Barah P, Jayavelu ND, Rasmussen S, Nielsen HB, Mundy J, Bones AM (2013) Genome-scale cold stress response regulatory networks in ten Arabidopsis thaliana ecotypes. BMC Genomics 14:5186–5190. doi:10.1186/1471-2164-14-722
Breusegem FV, Slooten L, Stassart JM, Botterman J, Moens T, Montagu MV, Inzé D (1999) Effects of overproduction of tobacco MnSOD in maize chloroplasts on foliar tolerance to cold and oxidative stress. J Exp Bot 50:71–78. doi:10.1093/jxb/50.330.71
Bu Q, Lv T, Shen H, Luong P, Wang J, Wang Z, Huang Z, Xiao L, Engineer C, Kim TH (2014) Regulation of drought tolerance by the F-box protein MAX2 in Arabidopsis. Plant Physiol 164:424–439. doi:10.1104/pp.113.226837
Castonguay Y, Laberge S, Brummer EC, Volenec JJ (2006) Alfalfa winter hardiness: a research retrospective and integrated perspective*. Adv Agron 90:203–265. doi:10.1016/S0065-2113(06)90006-6
Castonguay Y, Cloutier J, Bertrand A, Michaud R, Laberge S (2010) SRAP polymorphisms associated with superior freezing tolerance in alfalfa (Medicago sativa spp. sativa). Theor Appl Genet 120:1611–1619. doi:10.1007/s00122-010-1280-2
Chen THH, Murata N (2002) Enhancement of tolerance of abiotic stress by metabolic engineering of betaines and other compatible solutes. Curr Opin Plant Biol 5:250–257. doi:10.1016/S1369-5266(02)00255-8
Chen JR, Lü JJ, Liu R, Xiong XY, Wang T, Chen SY, Guo LB, Wang HF (2010) DREB1C from Medicago truncatula enhances freezing tolerance in transgenic M. truncatula and China rose (Rosa chinensis Jacq.). Plant Growth Regul 60:199–211. doi:10.1007/s10725-009-9434-4
Chen J, Tian Q, Pang T, Jiang L, Wu R, Xia X, Yin W (2014) Deep-sequencing transcriptome analysis of low temperature perception in a desert tree, Populus euphratica. BMC Genomics 15:326
Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17:1043–1054. doi:10.1101/gad.1077503
Chinnusamy V, Zhu J, Zhu J (2006) Gene regulation during cold acclimation in plants. Physiol Plant 126:52–61. doi:10.1111/j.1399-3054.2006.00596.x
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. doi:10.1093/bioinformatics/bti610
Cox SE, Stushnoff C (2001) Temperature-related shifts in soluble carbohydrate content during dormancy and cold acclimation in Populus tremuloides. Can J For Res 31:730–737. doi:10.1139/x00-206
Davey MP, Woodward FI, Quick WP (2009) Intraspecific variation in cold-temperature metabolic phenotypes of Arabidopsis lyrata ssp. petraea. Metabolomics 5:138–149. doi:10.1007/s11306-008-0127-1
Die JV, Rowland LJ (2014) Elucidating cold acclimation pathway in blueberry by transcriptome profiling. Environ Exp Bot 106:87–98. doi:10.1016/j.envexpbot.2013.12.017
Dong A, Yang J, Peng Z (2012) Transcriptome profiling of low temperature-treated cassava apical shoots showed dynamic responses of tropical plant to cold stress. BMC Genomics 13:64. doi:10.1186/1471-2164-13-64
Dong X, Im SB, Lim YP, Nou IS, Hur Y (2014) Comparative transcriptome profiling of freezing stress responsiveness in two contrasting Chinese cabbage genotypes, Chiifu and Kenshin. Genes Genomics 36:215–227. doi:10.1007/s13258-013-0160-y
Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38:64–70. doi:10.1093/nar/gkq310
Du H, Liu H, Xiong L (2013) Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front Plant Sci 4:397. doi:10.3389/fpls.2013.00397
Ecker JR (1995) The ethylene signal transduction pathway in plants. Science 268:667. doi:10.1007/978-94-011-4453-7_11
Ezaki B, Suzuki M, Motoda H, Kawamura M, Nakashima S, Matsumoto H (2004) Mechanism of gene expression of Arabidopsis glutathione S-transferase, AtGST1, and AtGST11 in response to aluminum stress. Plant Physiol 134:1672–1682. doi:10.1104/pp.103.037135
Fonseca S, Chico JM, Solano R (2009) The jasmonate pathway: the ligand, the receptor and the core signalling module. Curr Opin Plant Biol 12:539–547. doi:10.1016/j.pbi.2009.07.013
Goodwin W, Pallas JA, Jenkins GI (1996) Transcripts of a gene encoding a putative cell wall-plasma membrane linker protein are specifically cold-induced in Brassica napus. Plant Mol Biol 31:771–781. doi:10.1007/BF00019465
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Xian A, Fan L, Raychowdhury R, Zeng Q (2013) Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol 29. doi:10.1038/nbt.1883
Harberd N (2009) The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: how an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 21:1328–1339. doi:10.1105/tpc.109.066969
Hiroaki F, Viswanathan C, Americo R, Silvia R, Regina A, Sang-Youl P, Cutler SR, Jen S, Rodriguez PL, Jian-Kang Z (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462:660–664. doi:10.1038/nature08599
Horvath DP, Chao WS, Suttle JC, Thimmapuram J, Anderson JV (2008) Transcriptome analysis identifies novel responses and potential regulatory genes involved in seasonal dormancy transitions of leafy spurge (Euphorbia esula L.). BMC Genomics 9:536. doi:10.1186/1471-2164-9-536
Huang GQ, Gong SY, Xu WL, Li P, Zhang DJ (2011) GhHyPRP4, a cotton gene encoding putative hybrid proline-rich protein, is preferentially expressed in leaves and involved in plant response to cold stress. Acta Biochim Biophys Sin 43:519–527. doi:10.1093/abbs/gmr040
Hui YU, Yao JH, Liu R, Cui GW, Hou SQ (2010) Comprehensive evaluation on forage yield, nutrition quality and winter surviving rate of different alfalfa varieties. Chin J Grassland 32:108–111
Janmohammadi M (2013) Metabolomic analysis of low temperature responses in plants. Curr Opin Agric: 1
Janská A, Mar P, Zelenková S, Ovesná J (2010) Cold stress and acclimation—what is important for metabolic adjustment? Plant Biol 12:395–405. doi:10.1111/j.1438-8677.2009.00299.x
Janská A, Aprile A, Cattivelli L, Zámečník J, Bellis LD, Ovesná J (2014) The up-regulation of elongation factors in the barley leaf and the down-regulation of nucleosome assembly genes in the crown are both associated with the expression of frost tolerance. Funct Integr Genomics 14:493–506. doi:10.1007/s10142-014-0377-0
Jian BS, Yan XW, Hai BL, Bo WL, Zhao SZ, Shuai G, Zhi MY (2015) The F-box family genes as key elements in response to salt, heavy mental, and drought stresses in Medicago truncatula. Funct Integr Genomics 15:1–13. doi:10.1007/s10142-015-0438-z
Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, Guy CL (2004) Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol 136:4159–4168. doi:10.1104/pp.104.052142
Kawamura Y, Uemura M (2003) Mass spectrometric approach for identifying putative plasma membrane proteins of Arabidopsis leaves associated with cold acclimation. Plant J 36:141–154. doi:10.1046/j.1365-313X.2003.01864.x
Kazan K (2015) Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci 20:219–229. doi:10.1016/j.tplants.2015.02.001
Korn M, Gärtner T, Erban A, Kopka J, Selbig J, Hincha DK (2010) Predicting Arabidopsis freezing tolerance and heterosis in freezing tolerance from metabolite composition. Mol Plant 3:224–235. doi:10.1093/mp/ssp105
Li J, Tax FE (2013) Receptor-like kinases: key regulators of plant development and defense. J Integr Plant Biol 55:1184–1187. doi:10.1111/jipb.12129
Li ZY, Xing XF, Tang H, Yin YL, Guo YJ (2013) Effects of aluminum and acid stresses on the growth and antioxidant enzyme activities of rhizobia isolated from Medicago lupulina and M. sativa. Acta Prataculturae Sin 22:146–153
Liao P, Chen Q, Chye M (2014) Transgenic Arabidopsis flowers overexpressing acyl-CoA-binding protein ACBP6 are freezing tolerant. Plant Cell Physiol 55:1055–1071. doi:10.1093/pcp/pcu037
Liu H, Ouyang B, Zhang J, Wang T, Li H, Zhang Y, Yu C, Ye Z (2012) Differential modulation of photosynthesis, signaling, and transcriptional regulation between tolerant and sensitive tomato genotypes under cold stress. PLoS One 7. doi:10.1371/journal.pone.0050785
Liu A, Yu Y, Li R, Duan X, Zhu D, Sun X, Duanmu H, Zhu Y (2015) A novel hybrid proline-rich type gene GsEARLI17 from Glycine soja participated in leaf cuticle synthesis and plant tolerance to salt and alkali stresses. Plant Cell Tissue Organ Cult 121:633–646. doi:10.1007/s11240-015-0734-2
Lyzenga WJ, Stone SL (2012) Abiotic stress tolerance mediated by protein ubiquitination. J Exp Bot 63:599–616. doi:10.1093/jxb/err310
Mao XZ, Cai T, Olyarchuk JG, Wei LP (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21:3787–3793. doi:10.1093/bioinformatics/bti430
Maruyama K, Urano K, Yoshiwara K, Morishita Y, Sakurai N, Suzuki H, Kojima M, Sakakibara H, Shibata D, Saito K (2014) Integrated analysis of the effects of cold and dehydration on rice metabolites, phytohormones, and gene transcripts. Plant Physiol 164:1759–1771. doi:10.1104/pp.113.231720
Maurel C (2007) Plant aquaporins: novel functions and regulation properties. FEBS Lett 581:2227–2236. doi:10.1016/j.febslet.2007.03.021
Osakabe Y, Yamaguchi-Shinozaki K, Shinozaki K, Tran LSP (2014) ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol 202:35–49. doi:10.1111/nph.12613
Pang T, Ye CY, Xia XL, Yin WL (2013) De novo sequencing and transcriptome analysis of the desert shrub, Ammopiptanthus mongolicus, during cold acclimation using Illumina/Solexa. BMC Genomics 14:488. doi:10.1186/1471-2164-14-488
Pembleton KG, Volenec JJ, Rawnsley RP, Donaghy DJ (2010) Partitioning of taproot constituents and crown bud development are affected by water deficit in regrowing alfalfa (L.). Crop Sci 50:989–999. doi:10.2135/cropsci2009.03.0140
Peng Y, Lin W, Cai W, Arora R (2007) Overexpression of a Panax ginseng tonoplast aquaporin alters salt tolerance, drought tolerance and cold acclimation ability in transgenic Arabidopsis plants. Planta 226:729–740. doi:10.1007/s00425-007-0520-4
Pérez-Rodríguez P, Riano-Pachon DM, Corrêa LGG, Rensing SA, Kersten B, Mueller-Roeber B (2010) PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res 38:822–827. doi:10.1093/nar/gkp805
Postnikova OA, Shao J, Nemchinov LG (2013) Analysis of the alfalfa root transcriptome in response to salinity stress. Plant Cell Physiol 54:1041–1055. doi:10.1093/pcp/pct056
Priyanka B, Sekhar K, Reddy VD, Rao KV (2010) Expression of pigeonpea hybrid-proline-rich protein encoding gene (CcHyPRP) in yeast and Arabidopsis affords multiple abiotic stress tolerance: pigeonpea gene confers multiple stress tolerance. Plant Biotechnol J 8:76–87. doi:10.1111/j.1467-7652.2009.00467.x
Qin LX, Zhang DJ, Huang GQ, Li L, Li J, Gong SY, Li XB, Xu WL (2013) Cotton GhHyPRP3 encoding a hybrid proline-rich protein is stress inducible and its overexpression in Arabidopsis enhances germination under cold temperature and high salinity stress conditions. Acta Physiol Plant 35:1531–1542. doi:10.1007/s11738-012-1194-5
Rasmussen S, Cao M, Fraser K, Koulman A, Park-Ng Z, Xue H, Lane G (2006) Cold stress in white clover—an integrated view of metabolome and transcriptome responses. Breeding for success: diversity in action. Proceedings of the 13th Australasian Plant Breeding Conference, Christchurch, New Zealand, pp 750–757
Rinne PLH, Kaikuranta PM, Christiaan VDS (2001) The shoot apical meristem restores its symplasmic organization during chilling-induced release from dormancy. Plant J 26:249–264. doi:10.1046/j.1365-313X.2001.01022.x
Rohde A, Ruttink T, Hostyn V, Sterck L, Van Driessche K, Boerjan W (2007) Gene expression during the induction, maintenance, and release of dormancy in apical buds of poplar. J Exp Bot 58:4047–4060. doi:10.1093/jxb/erm261
Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration and cold-inducible gene expression. Biochem Biophys Res Commun 290:998–1009. doi:10.1006/bbrc.2001.6299
Schwab PM, Barnes DK, Sheaffer CC (1996) The relationship between field winter injury and fall growth score for 251 alfalfa cultivars. Cropence 36:418–426. doi:10.2135/cropsci1996.0011183X003600020034x
Song YP, Chen QQ, Dong C, Zhang DQ (2013) Transcriptome profiling reveals differential transcript abundance in response to chilling stress in Populus simonii. Plant Cell Rep 32:1407–1425. doi:10.1007/s00299-013-1454-x
Song Y, Dong C, Min T, Zhang D (2014) Comparison of the physiological effects and transcriptome responses of Populus simonii under different abiotic stresses. Plant Mol Biol 86:139–156. doi:10.1007/s11103-014-0218-5
Stitt M, Hurry V (2002) A plant for all seasons: alterations in photosynthetic carbon metabolism during cold acclimation in Arabidopsis. Curr Opin Plant Biol 5:199–206
Sun QZ, Wang YQ, Hou XY (2004) Alfalfa winter survival research summary. Pratacult Sci 21:21–25
Suzuki N, Mittler R (2006) Reactive oxygen species and temperature stresses: a delicate balance between signaling and destruction. Physiol Plant 126:45–51. doi:10.1111/j.0031-9317.2005.00582.x
Svensson JT, Crosatti C, Campoli C, Bassi R, Stanca AM, Close TJ, Cattivelli L (2006) Transcriptome analysis of cold acclimation in barley Albina and Xantha mutants. Plant Physiol 141:257–270. doi:10.1104/pp.105.072645
Tayeh N, Bahrman N, Sellier H, Bluteau A, Blassiau C, Fourment J, Bellec A, Debellé F, Lejeune-Hénaut I, Delbreil B (2013) A tandem array of CBF/DREB1 genes is located in a major freezing tolerance QTL region on Medicago truncatula chromosome 6. BMC Genomics 14:814–830. doi:10.1186/1471-2164-14-814
Thomashow MF (1999) PLANT COLD ACCLIMATION: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50:571–599. doi:10.1146/annurev.arplant.50.1.571
Tommasini L, Svensson JT, Rodriguez EM, Wahid A, Malatrasi M, Kato K, Wanamaker S, Resnik J, Close TJ (2008) Dehydrin gene expression provides an indicator of low temperature and drought stress: transcriptome-based analysis of Barley (Hordeum vulgare L.). Funct Integr Genom 8:387–405. doi:10.1007/s10142-008-0081-z
Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci 106:17588–17593. doi:10.1073/pnas.0907095106
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40:115–126. doi:10.1093/nar/gks596
Vierstra RD (1987) Ubiquitin, a key component in the degradation of plant proteins. Physiol Plant 70:103–106. doi:10.1111/j.1399-3054.1987.tb08704.x
Wang XC, Zhao QY, Ma CL, Zhang ZH, Cao HL, Kong YM, Yue C, Hao XY, Chen L, Ma JQ (2013a) Global transcriptome profiles of Camellia sinensis during cold acclimation. BMC Genomics 14:1–15. doi:10.1186/1471-2164-14-415
Wang H, Zou Z, Wang S, Gong M (2013a) Global analysis of transcriptome responses and gene expression profiles to cold stress of Jatropha curcas L. PLoS One 8. doi:10.1371/journal.pone.0082817
Wang C, Gao CQ, Wang LQ, Zheng L, Yang CP, Wang YC (2014) Comprehensive transcriptional profiling of NaHCO3-stressed Tamarix hispida roots reveals networks of responsive genes. Plant Mol Biol 84:145–157. doi:10.1007/s11103-013-0124-2
Wei S, Du Z, Gao F, Ke X, Li J, Liu J, Zhou Y (2015) Global transcriptome profiles of ‘Meyer’ Zoysiagrass in response to cold stress. PLoS One 10. doi:10.1371/journal.pone.0131153
Winfield MO, Lu CG, Wilson ID, Coghill JA, Edwards KJ (2010) Plant responses to cold: transcriptome analysis of wheat. Plant Biotechnol J 8:749–771. doi:10.1111/j.1467-7652.2010.00536.x
Wu Y, Wei W, Pang X, Wang X, Zhang H, Bo D, Xing Y, Li X, Wang M (2014) Comparative transcriptome profiling of a desert evergreen shrub, Ammopiptanthus mongolicus, in response to drought and cold stresses. BMC Genomics 15:1–16. doi:10.1186/1471-2164-15-671
Wu ZJ, Li XH, Liu ZW, Li H, Wang YX, Zhuang J (2015) Transcriptome-based discovery of AP2/ERF transcription factors related to temperature stress in tea plant (Camellia sinensis). Funct Integr Genomics 15:741–752. doi:10.1007/s10142-015-0457-9
Xu J, Tian YS, Xing XJ, Peng RH, Zhu B, Gao JJ, Yao QH (2016) Over-expression of AtGSTU19 provides tolerance to salt, drought and methyl viologen stresses in Arabidopsis. Physiol Plant 156:164–175. doi:10.1111/ppl.12347
Yan YS, Chen XY, Yang K, Sun ZX, Fu YP, Zhang YM, Fang RX (2011) Overexpression of an F-box protein gene reduces abiotic stress tolerance and promotes root growth in rice. Mol Plant 4:190–197. doi:10.1093/mp/ssq066
Yang SS, Tu ZJ, Cheung F, Xu WW, Lamb JAF, Jung HJG, Vance CP, Gronwald JW (2011) Using RNA-Seq for gene identification, polymorphism detection and transcript profiling in two alfalfa genotypes with divergent cell wall composition in stems. BMC Genomics 12:199–218. doi:10.1186/1471-2164-12-199
Ye J, Fang L, Zheng HK, Zhang Y, Chen J, Zhang ZJ, Wang J, Li ST, Li RQ, Bolund L, Wang J (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34:293–297. doi:10.1093/nar/gkl031
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14–R25. doi:10.1186/gb-2010-11-2-r14
Yu X, Pijut PM, Byrne S, Asp T, Bai G, Jiang Y (2015) Candidate gene association mapping for winter survival and spring regrowth in perennial ryegrass. Plant Sci 235:37–45. doi:10.1016/j.plantsci.2015.03.003
Zhang CK, Lang P, Dane F, Ebel RC, Singh NK, Locy RD, William AD (2005) Cold acclimation induced genes of trifoliate orange (Poncirus trifoliata). Plant Cell Rep 23:764–769. doi:10.1007/s00299-004-0883-y
Zhang LL, Zhao MG, Tian QY, Zhang WH (2011) Comparative studies on tolerance of Medicago truncatula and Medicago falcata to freezing. Planta 234:445–457
Zhang XJ, Yang GY, Shi R, Han XM, Qi LW, Wang RG, Xiong LM, Li GJ (2013) Arabidopsis cysteine-rich receptor-like kinase 45 functions in the responses to abscisic acid and abiotic stresses. Plant Physiol Biochem 67:189–198. doi:10.1016/j.plaphy.2013.03.013
Zhang JY, Cruz de Carvalho MH, Torres-Jerez I, Kang Y, Allen SN, Huhman DV, Tang YH, Murray J, Sumner LW, Udvardi MK (2014) Global reprogramming of transcription and metabolism in Medicago truncatula during progressive drought and after rewatering. Plant Cell Environ 37:2553–2576. doi:10.1111/pce.12328
Zhang SH, Shi YH, Cheng NN, Du HQ, Fan WN, Wang CZ (2015) De novo characterization of fall dormant and nondormant alfalfa (Medicago sativa L.) leaf transcriptome and identification of candidate genes related to fall dormancy. PLoS One 10. doi:10.1371/journal.pone.0122170
Zhou S, Sun X, Yin S, Kong X, Shan Z, Ying X, Yin L, Wei W (2014) The role of the F-box gene TaFBA1 from wheat (Triticum aestivum L.) in drought tolerance. Plant Physiol Biochem 84c:213–223. doi:10.1016/j.plaphy.2014.09.017
Zhou SM, Kong XZ, Kang HH, Sun XD, Wang W (2015) The involvement of wheat F-box protein gene TaFBA1 in the oxidative stress tolerance of plants. PLoS One 10. doi:10.1371/journal.pone.0122117
Zhu J, Dong CH, Zhu JK (2007) Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr Opin Plant Biol 10:290–295. doi:10.1016/j.pbi.2007.04.010
Acknowledgments
This work was supported by the MOST 863 project (2013AA102607-5), the Graduate Innovation Fund of Harbin Normal University (HSDBSCX2014-04), Key Scientific and Technological Project of Heilongjiang Province of China (GA15B105-1), and the Natural and Science Foundation of China (Nos. 31302019 and 31470571).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Song, L., Jiang, L., Chen, Y. et al. Deep-sequencing transcriptome analysis of field-grown Medicago sativa L. crown buds acclimated to freezing stress. Funct Integr Genomics 16, 495–511 (2016). https://doi.org/10.1007/s10142-016-0500-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-016-0500-5