Abstract
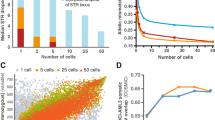
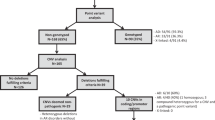
Different types of large NF1 deletion are distinguishable by breakpoint location and potentially also by the frequency of mosaicism with normal cells lacking the deletion. However, low-grade mosaicism with fewer than 10% normal cells has not yet been excluded for all NF1 deletion types since it is impossible to assess by the standard techniques used to identify such deletions, including MLPA and array analysis. Here, we used ultra-deep amplicon sequencing to investigate the presence of normal cells in the blood of 20 patients with type-1 NF1 deletions lacking mosaicism according to MLPA. The ultra-deep sequencing entailed the screening of 96 amplicons for heterozygous SNVs located within the NF1 deletion region. DNA samples from three previously identified patients with type-2 NF1 deletions and low-grade mosaicism with normal cells as determined by FISH or microsatellite marker analysis were used to validate our methodology. In these type-2 NF1 deletion samples, proportions of 5.3%, 6.6% and 15.0% normal cells, respectively, were detected by ultra-deep amplicon sequencing. However, using this highly sensitive method, none of the 20 patients with type-1 NF1 deletions included in our analysis exhibited low-grade mosaicism with normal cells in blood, thereby supporting the view that the vast majority of type-1 deletions are germline deletions.

Similar content being viewed by others
References
Alhourani E, Rincic M, Othman MAK, Pohle B, Schlie C, Glaeser A, Liehr T (2014) Comprehensive chronic lymphocytic leukemia diagnostics by combined multiplex ligation dependent probe amplification (MLPA) and interphase fluorescence in situ hybridization (iFISH). Mol Cytogenet 7:79
Bengesser K, Cooper DN, Steinmann K, Kluwe L, Chuzhanova NA, Wimmer K, Tatagiba M, Tinschert S, Mautner VF, Kehrer-Sawatzki H (2010) A novel third type of recurrent NF1 microdeletion mediated by nonallelic homologous recombination between LRRC37B-containing low-copy repeats in 17q11.2. Hum Mutat 31:742–751
Bengesser K, Vogt J, Mussotter T, Mautner VF, Messiaen L, Cooper DN, Kehrer-Sawatzki H (2014) Analysis of crossover breakpoints yields new insights into the nature of the gene conversion events associated with large NF1 deletions mediated by nonallelic homologous recombination. Hum Mutat 35:215–226
Bernkopf M, Hunt D, Koelling N, Morgan T, Collins AL, Fairhurst J, Robertson SP, Douglas AGL, Goriely A (2017) Quantification of transmission risk in a male patient with a FLNB mosaic mutation causing Larsen syndrome: Implications for genetic counseling in postzygotic mosaicism cases. Hum Mutat 38:1360–1364
Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K (2006) Genome-wide mapping of polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20:1123–1136
Brodin J, Mild M, Hedskog C, Sherwood E, Leitner T, Andersson B, Albert J (2013) PCR-induced transitions are the major source of error in cleaned ultra-deep pyrosequencing data. PLoS One 8:e70388
Chang F, Liu L, Fang E, Zhang G, Chen T, Cao K, Li Y, Li MM (2017) Molecular diagnosis of mosaic overgrowth syndromes using a custom-designed next-generation sequencing panel. J Mol Diagn 19:613–624
Cnossen MH, van der Est MN, Breuning MH, van Asperen CJ, Breslau-Siderius EJ, van der Ploeg AT, de Goede-Bolder A, van den Ouweland AM, Halley DJ, Niermeijer MF (1997) Deletions spanning the neurofibromatosis type 1 gene: implications for genotype-phenotype correlations in neurofibromatosis type 1? Hum Mutat 9:458–464
Cohen AS, Wilson SL, Trinh J, Ye XC (2015) Detecting somatic mosaicism: considerations and clinical implications. Clin Genet 87:554–562
Coll-Mulet L, Santidrián AF, Cosialls AM, Iglesias-Serret D, de Frias M, Grau J, Menoyo A, González-Barca E, Pons G, Domingo A, Gil J (2008) Multiplex ligation-dependent probe amplification for detection of genomic alterations in chronic lymphocytic leukaemia. Br J Haematol 142:793–801
Contini E, Paganini I, Sestini R, Candita L, Capone GL, Barbetti L, Falconi S, Frusconi S, Giotti I, Giuliani C, Torricelli F, Benelli M, Papi L (2015) A systematic assessment of accuracy in detecting somatic mosaic variants by deep amplicon sequencing: application to NF2 gene. PLoS One 10:e0129099
Davidson CJ, Zering E, Champion KJ, Gauthier MP, Wang F, Boonyaratanakornkit J, Jones JR, Schreiber E (2012) Improving the limit of detection for Sanger sequencing: a comparison of methodologies for KRAS variant detection. Biotechniques 53:182–188
De Raedt T, Stephens M, Heyns I, Brems H, Thijs D, Messiaen L, Stephens K, Lazaro C, Wimmer K, Kehrer-Sawatzki H, Vidaud D, Kluwe L, Marynen P, Legius E (2006) Conservation of hotspots for recombination in low-copy repeats associated with the NF1 microdeletion. Nat Genet 38:1419–1423
Dorschner MO, Sybert VP, Weaver M, Pletcher BA, Stephens K (2000) NF1 microdeletion breakpoints are clustered at flanking repetitive sequences. Hum Mol Genet 9:35–46
Forbes SH, Dorschner MO, Le R, Stephens K (2004) Genomic context of paralogous recombination hotspots mediating recurrent NF1 region microdeletion. Genes Chromosomes Cancer 41:12–25
Gajecka M (2016) Unrevealed mosaicism in the next-generation sequencing era. Mol Genet Genomics 291:513–530
Gottipati P, Helleday T (2009) Transcription-associated recombination in eukaryotes: link between transcription, replication and recombination. Mutagenesis 24:203–210
Hillmer M, Wagner D, Summerer A, Daiber M, Mautner VF, Messiaen L, Cooper DN, Kehrer-Sawatzki H (2016) Fine mapping of meiotic NAHR-associated crossovers causing large NF1 deletions. Hum Mol Genet 25:484–496
Hillmer M, Summerer A, Mautner VF, Högel J, Cooper DN, Kehrer-Sawatzki H (2017) Consideration of the haplotype diversity at nonallelic homologous recombination hotspots improves the precision of rearrangement breakpoint identification. Hum Mutat 38:1711–1722
Hömig-Hölzel C, Savola S (2012) Multiplex ligation-dependent probe amplification (MLPA) in tumor diagnostics and prognostics. Diagn Mol Pathol 21:189–206
Jenne DE, Tinschert S, Reimann H, Lasinger W, Thiel G, Hameister H, Kehrer-Sawatzki H (2001) Molecular characterization and gene content of breakpoint boundaries in patients with neurofibromatosis type 1 with 17q11.2 microdeletions. Am J Hum Genet 69:516–527
Kehrer-Sawatzki H, Cooper DN (2008) Mosaicism in sporadic neurofibromatosis type 1: variations on a theme common to other hereditary cancer syndromes? J Med Genet 45:622–631
Kehrer-Sawatzki H, Kluwe L, Sandig C, Kohn M, Wimmer K, Krammer U, Peyrl A, Jenne DE, Hansmann I, Mautner VF (2004) High frequency of mosaicism among patients with neurofibromatosis type 1 (NF1) with microdeletions caused by somatic recombination of the JJAZ1 gene. Am J Hum Genet 75:410–423
Kehrer-Sawatzki H, Mautner VF, Cooper DN (2017) Emerging genotype-phenotype relationships in patients with large NF1 deletions. Hum Genet 136:349–376
Kluwe L, Siebert R, Gesk S, Friedrich RE, Tinschert S, Kehrer-Sawatzki H, Mautner VF (2004) Screening 500 unselected neurofibromatosis 1 patients for deletions of the NF1 gene. Hum Mutat 23:111–116
Kumar D, Mehta A, Panigrahi MK, Nath S, Saika KK (2018) NPM1 mutation analysis in acute myeloid leukemia: comparison of three techniques—Sanger sequencing, pyrosequencing, and real-time polymerase chain reaction. Turk J Haematol 35:49–53
López-Correa C, Brems H, Lázaro C, Marynen P, Legius E (2000) Unequal meiotic crossover: a frequent cause of NF1 microdeletions. Am J Hum Genet 66:1969–1974
López-Correa C, Dorschner M, Brems H, Lázaro C, Clementi M, Upadhyaya M, Dooijes D, Moog U, Kehrer-Sawatzki H, Rutkowski JL, Fryns JP, Marynen P, Stephens K, Legius E (2001) Recombination hotspot in NF1 microdeletion patients. Hum Mol Genet 10:1387–1392
Marnef A, Cohen S, Legube G (2017) Transcription-coupled DNA double-strand break repair: active genes need special care. J Mol Biol 429:1277–1288
Mautner VF, Kluwe L, Friedrich RE, Roehl AC, Bammert S, Högel J, Spöri H, Cooper DN, Kehrer-Sawatzki H (2010) Clinical characterisation of 29 neurofibromatosis type-1 patients with molecularly ascertained 1.4 Mb type-1 NF1 deletions. J Med Genet 47:623–630
Messiaen L, Vogt J, Bengesser K, Fu C, Mikhail F, Serra E, Garcia-Linares C, Cooper DN, Lazaro C, Kehrer-Sawatzki H (2011) Mosaic type-1 NF1 microdeletions as a cause of both generalized and segmental neurofibromatosis type-1 (NF1). Hum Mutat 32:213–219
Morimoto Y, Ono S, Imamura A, Okazaki Y, Kinoshita A, Mishima H, Nakane H, Ozawa H, Yoshiura KI, Kurotaki N (2017) Deep sequencing reveals variations in somatic cell mosaic mutations between monozygotic twins with discordant psychiatric disease. Hum Genome Var 4:17032
Neuhäusler L, Summerer A, Cooper DN, Mautner VF, Kehrer-Sawatzki H (2018) Pronounced maternal parent-of-origin bias for type-1 NF1 microdeletions. Hum Genet 137:365–373
Oneda B, Asadollahi R, Azzarello-Burri S, Niedrist D, Baldinger R, Masood R, Schinzel A, Latal B, Jenni OG, Rauch A (2017) Low-level chromosomal mosaicism in neurodevelopmental disorders. Mol Syndromol 8:266–271
Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K (2007) The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol 27:3769–3779
Pasmant E, Sabbagh A, Spurlock G, Laurendeau I, Grillo E, Hamel MJ, Martin L, Barbarot S, Leheup B, Rodriguez D, Lacombe D, Dollfus H, Pasquier L, Isidor B, Ferkal S, Soulier J, Sanson M, Dieux-Coeslier A, Bièche I, Parfait B, Vidaud M, Wolkenstein P, Upadhyaya M, Vidaud D, Members of the NF France Network (2010) NF1 microdeletions in neurofibromatosis type 1: from genotype to phenotype. Hum Mutat 31:E1506–e1518
Qin L, Wang J, Tian X, Yu H, Truong C, Mitchell JJ, Wierenga KJ, Craigen WJ, Zhang VW, Wong LC (2016) Detection and quantification of mosaic mutations in disease genes by next-generation sequencing. J Mol Diagn 18:446–453
Rasmussen SA, Colman SD, Ho VT, Abernathy CR, Arn PH, Weiss L, Schwartz C, Saul RA, Wallace MR (1998) Constitutional and mosaic large NF1 gene deletions in neurofibromatosis type 1. J Med Genet 35:468–471
Roehl AC, Vogt J, Mussotter T, Zickler AN, Spöti H, Högel J, Chuzhanova NA, Wimmer K, Kluwe L, Mautner VF, Cooper DN, Kehrer-Sawatzki H (2010a) Intrachromosomal mitotic nonallelic homologous recombination is the major molecular mechanism underlying type-2 NF1 deletions. Hum Mutat 31:1163–1173
Roehl AC, Cooper DN, Kluwe L, Helbrich A, Wimmer K, Högel J, Mautner VF, Kehrer-Sawatzki H (2010b) Extended runs of homozygosity at 17q11.2: an association with type-2 NF1 deletions? Hum Mutat 31:325–334
Roehl AC, Mussotter T, Cooper DN, Kluwe L, Wimmer K, Högel J, Zetzmann M, Vogt J, Mautner VF, Kehrer-Sawatzki H (2012) Tissue-specific differences in the proportion of mosaic large NF1 deletions are suggestive of a selective growth advantage of hematopoietic del(+/−) stem cells. Hum Mutat 33:541–550
Shao W, Boltz VF, Spindler JE, Kearney MF, Maldarelli F, Mellors JW, Stewart C, Volfovsky N, Levitsky A, Stephens RM, Coffin JM (2013) Analysis of 454 sequencing error rate, error sources, and artifact recombination for detection of low-frequency drug resistance mutations in HIV-1 DNA. Retrovirology 10:18
Steinmann K, Cooper DN, Kluwe L, Chuzhanova NA, Senger C, Serra E, Lazaro C, Gilaberte M, Wimmer K, Mautner VF, Kehrer-Sawatzki H (2007) Type 2 NF1 deletions are highly unusual by virtue of the absence of nonallelic homologous recombination hotspots and an apparent preference for female mitotic recombination. Am J Hum Genet 81:1201–1220
Stevens-Kroef M, Simons A, Gorissen H, Feuth T, Weghuis DO, Buijs A, Raymakers R, Geurts van Kessel A (2009) Identification of chromosomal abnormalities relevant to prognosis in chronic lymphocytic leukemia using multiplex ligation-dependent probe amplification. Cancer Genet Cytogenet 195:97–104
Summerer A, Mautner VF, Upadhyaya M, Claes KBM, Högel J, Cooper DN, Messiaen L, Kehrer-Sawatzki H (2018) Extreme clustering of type-1 NF1 deletion breakpoints co-locating with G-quadruplex forming sequences. Hum Genet 137:511–520
Tsiatis AC, Norris-Kirby A, Rich RG, Hafez MJ, Gocke CD, Eshleman JR, Murphy KM (2010) Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. J Mol Diagn 12:425–432
van Veghel-Plandsoen MM, Wouters CH, Kromosoeto JN, den Ridder-Klünnen MC, Halley DJ, van den Ouweland AM (2011) Multiplex ligation-depending probe amplification is not suitable for detection of low-grade mosaicism. Eur J Hum Genet 19:1009–1012
Vogt J, Mussotter T, Bengesser K, Claes K, Högel J, Chuzhanova N, Fu C, van den Ende J, Mautner VF, Cooper DN, Messiaen L, Kehrer-Sawatzki H (2012) Identification of recurrent type-2 NF1 microdeletions reveals a mitotic nonallelic homologous recombination hotspot underlying a human genomic disorder. Hum Mutat 33:1599–1609
Vogt J, Bengesser K, Claes KB, Wimmer K, Mautner VF, van Minkelen R, Legius E, Brems H, Upadhyaya M, Högel J, Lazaro C, Rosenbaum T, Bammert S, Messiaen L, Cooper DN, Kehrer-Sawatzki H (2014) SVA retrotransposon insertion-associated deletion represents a novel mutational mechanism underlying large genomic copy number changes with non-recurrent breakpoints. Genome Biol 15:R80
Zhang J, Tong H, Fu X, Zhang Y, Liu J, Cheng R, Liang J, Peng J, Sun Z, Liu H, Zhang F, Lu W, Li M, Yao Z (2015) Molecular characterization of NF1 and neurofibromatosis type 1 genotype-phenotype correlations in a Chinese population. Sci Rep 5:11291
Zickler AM, Hampp S, Messiaen L, Bengesser K, Mussotter T, Roehl AC, Wimmer K, Mautner VF, Kluwe L, Upadhyaya M, Pasmant E, Chuzhanova N, Kestler HA, Högel J, Legius E, Claes K, Cooper DN, Kehrer-Sawatzki H (2012) Characterization of the nonallelic homologous recombination hotspot PRS3 associated with type-3 NF1 deletions. Hum Mutat 33:372–383
Acknowledgements
This work has been funded by the Deutsche Forschungsgemeinschaft (DFG) grant KE 724/12-2.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors are unaware of any conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Summerer, A., Schäfer, E., Mautner, VF. et al. Ultra-deep amplicon sequencing indicates absence of low-grade mosaicism with normal cells in patients with type-1 NF1 deletions. Hum Genet 138, 73–81 (2019). https://doi.org/10.1007/s00439-018-1961-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-018-1961-5