Abstract
The celiac disease (CD) is an inflammatory condition characterized by injury to the lining of the small-intestine on exposure to the gluten of wheat, barley and rye. The involvement of gluten in the CD syndrome has been studied in detail in bread wheat, where a set of “toxic” and “immunogenic” peptides has been defined. For wheat diploid species, information on CD epitopes is poor. In the present paper, we have adopted a genomic approach in order to understand the potential CD danger represented by storage proteins in diploid wheat and sequenced a sufficiently large number of cDNA clones related to storage protein genes of Triticum monococcum. Four bona fide toxic peptides and 13 immunogenic peptides were found. All the classes of storage proteins were shown to contain harmful sequences. The major conclusion is that einkorn has the full potential to induce the CD syndrome, as already evident for polyploid wheats. In addition, a complete overview of the storage protein gene arsenal in T. monococcum is provided, including a full-length HMW x-type sequence and two partial HMW y-type sequences.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The celiac disease (CD) is an inflammatory condition characterized by injury to the lining of the small-intestine on exposure to the gluten—specifically to its prolamin components—of wheat, barley and rye. The prevalence of CD among Caucasian is in a range 1:100–300 (Wieser and Koehler 2008). The disease shows a strong association with human leukocyte antigen (HLA) class II molecules DQ2 and DQ8, and CD4+ T cells specific for wheat gluten play a major role. The binding of gluten-derived peptides to the HLA molecules is enhanced when gluten is modified by tissue transglutaminase (tTG) that converts specific glutamine residues to negatively charged glutamic acid (for comprehensive recent reviews of CD see Kagnoff 2007 and Wieser and Koehler 2008).
A variety of in vivo, ex vivo and in vitro methods have been exploited to search for gluten peptides involved in the disease (Wieser 2004). The causal agent resides mainly in the gliadin fraction of gluten: all three main structural types of gliadins, α/β-, γ- and ω-gliadins are active (Howdle et al. 1984; Fluge et al. 1994). Nevertheless, in vivo and in vitro tests revealed that also the glutenin components can exacerbate CD (Vader et al. 2002; Molberg et al. 2003; Dewar et al. 2006).
Gluten is a cohesive mass present as a network in dough, to which it confers visco-elastic properties. It is mainly composed of the prolamins and its visco-elastic properties strongly rely upon the ratio of monomeric to polymeric protein aggregates and on the size distribution of polymers (Wrigley et al. 2006).
The prolamins are assigned to three groups: sulphur-poor (S-poor), S-rich and high molecular weight (HMW) prolamins (Shewry and Halford 2002). The S-poor prolamins consist essentially of ω-gliadins, account for about 11% of total storage proteins and contain little or no cysteine residues. They are predominantly monomeric, with Mr ranging from 30,000 to 80,000 Da, comprising a single domain made up almost entirely of a single repeat motif. A group of S-poor prolamins may associate to disulphide-bonded glutenin polymers, behaving as low molecular weight glutenin subunits (LMW-GS); they are known as D-type LMW glutenins and are considered mutant ω-gliadins in which the presence of single cysteine residues allows cross-linking (Masci et al. 1993).
The S-rich prolamins, accounting for about 70–80% of the prolamin fraction, have Mr from about 30,000 to 55,000 Da and include both monomeric α/β- and γ-gliadins, and polymeric LMW glutenins. They consist of a repetitive N-terminal domain, representing up to half of the molecule, and a non-repetitive cysteine rich C-terminal domain. In addition, α/β- and γ-gliadins contain two and one polyglutamine (polyGln) regions, and six and eight conserved cysteine residues, respectively. Cysteins (Cys) form either three or four intra-chain disulphide bonds; additional Cys can be present allowing the incorporation of α/β- and γ-gliadins into gluten polymers as bound gliadins. LMW glutenins have been assigned to the groups B, C and D (Jackson et al. 1983) on the basis of their mobility on SDS-PAGE. The D-type belongs to the S-poor prolamin fraction. The B-type has been divided into subgroups LMW-m, LMW-s and LMW-i, based on the first amino acid in the N-terminal sequence (Lew et al. 1992). C-type LMW seem closer to α/β- and γ-gliadins rather than to the B-type of LMW glutenins (Masci et al. 2002). The typical LMW-GS contain a polyGln region and six conserved cysteine residues, forming three intra-chain disulphide bonds, plus one or more additional Cys contributing to the formation of inter-chain bonds (for a detailed review on LMW-GS see D’Ovidio and Masci 2004).
The HMW prolamins consist of HMW glutenins and constitute 10% of the prolamin fraction. They can be grouped into x- and y-type subunits, with Mr ranging from 83,000 to 88,000 Da and 67,000 to 74,000 Da, respectively. The HMW molecule comprises three structural domains, i.e. a central repetitive domain flanked by non-repetitive N- and C-terminal domains. Their molecules exist only as a component of high molecular weight polymers stabilised by inter-chain disulphide bonds (Shewry et al. 1992).
A resurging interest on cereals, and specifically wheat storage proteins, derives from the current advancement in the identification of putative epitopes responsible for the celiac disease syndrome. Such epitopes consist of peptides present in prolamin molecules. Sturgess et al. (1994) provided in vivo evidence for activity of a peptide corresponding to amino acid residues 31–49 (LGQQQPFPPQQPYPQPQPF) of a α/β-gliadin. Further results revealed that the response to gluten focuses on more than one gluten epitope, mainly belonging to α/β- and γ-gliadins: Arentz-Hansen et al. (2000) demonstrated that two overlapping peptides spanning the region 57–75 of α/β-gliadins (α9, 57QLQPFPQPQLPY68 and α2, 62PQPQLPYPQPQLPY75) are relevant when a single glutamine residue is deamidated to glutamic acid by tTG. The same authors (Arentz-Hansen et al. 2002) reported an additional α/β- and several γ-gliadin epitopes located in Proline-rich regions and provided evidence that deamidation is not an absolute requirement for T-cell activation in the very early stages of the disease. This is in agreement with Vader et al. (2002): the immune response was more heterogeneous in children than in adults. Moreover, a broad group of native gluten peptides, derived from both gliadins and glutenins, was found to activate CD in children, but when age increased the repertoire became narrow and focused on few immuno-dominant peptides capable of stronger binding affinity to DQ2 molecules. Deamidation of specific glutamine residues by tTG released from cytoplasmic stores, as a consequence of increasing tissue damage, may probably be the key step of epitope focusing. They also demonstrated that three out of the six novel gluten peptides, one spanning region 93–106 of α/β-gliadins (Glia-α20, PFRPQQPYPQPQPQ), one region 222–236 of γ-gliadins (Glia-γ30, VQGQIIQPQQPAQL) and a sequence (Glu-5, QQXSQPQXPQQQQXPQQPQQF, where X is Ile or Leu) that does not exactly match any of the protein in the database but contains the minimal epitope QXPQQPQQF, were also able to induce T-cell responses in adult CD patients. Van de Wal et al. (1998) reported the identification of a stimulating epitope residing in the C-terminal region of a α/β-gliadin (205PSGQGSFQPSQQ216). Data available concur to define a kind of epitope clustering in both α/β- and γ-gliadins, interestingly always included in proline-rich regions (Arentz-Hansen et al. 2002).
An understanding of the potential CD danger represented by wheat storage proteins can be obtained by (1) following the genetics of the mentioned protein products, with the aim of defining rules and location of the responsible loci; and (2) adopting a genomic approach.
The first line of studies has a long tradition (for a comprehensive review on the genetics of gluten proteins see Shewry et al. 2003). Most of the prolamins are encoded by multigene families located at complex loci mainly on group 1 and 6 chromosomes. S-poor prolamins (ω-gliadins, D-type LMW glutenins) are coded by the Gli-1 loci on the short arm of group 1 chromosomes; minor components are encoded by linked additional genes. The gene copy number is estimated to be 15–18 in hexaploid bread wheat. The S-rich prolamins (α/β- and γ-gliadins, and polymeric LMW glutenins) are encoded by three major series of homoeologous loci: Gli-1, responsible for the synthesis of γ-gliadins, Glu-3, tightly linked to Gli-1, encoding the B group of LMW glutenins, and Gli-2, on the short arm of chromosome group 6, responsible for α-type gliadins. Estimates of gene copy numbers vary from 17 to 39 for γ-gliadins, to 22–39 for LMW glutenins, to 60–150 for the α-type gliadins.
HMW prolamins are encoded by three Glu-1 loci on the long arm of chromosomes 1A, 1B and 1D of hexaploid bread wheat (Glu-A1, Glu-B1, Glu-D1). Each homoeologous locus contains two closely linked HMW genes encoding for a x-type subunit of higher Mr (x-type HMW) and a y-type subunit of lower Mr (y-type HMW). However, only three to five HMW subunits (out of six) are expressed in hexaploid bread wheat due to silencing of HMW subunit genes on 1A and 1B (Payne 1987). In durum wheats (T. durum) one to three (out of four) HMW subunits (Branlard et al. 1989) and in diploid wheats one or two (out of two) subunits (Waines and Payne 1987) are expressed. Allelic variation has been reported in the subunits encoded by each Glu-1 locus mainly in bread wheat but only few full-length HMW sequences have been reported so far (Shewry et al. 2003).
For einkorn, T. monococcum, we adopted the genomic approach, which consists in sequencing a sufficiently large number of cDNA clones related to seed storage proteins. The scope of this paper is therefore to define the number of genes encoding CD epitopes in this species, including a complete overview of its storage protein gene arsenal.
Materials and methods
cDNA library construction from T. monococcum grains
Poly A+ RNA isolation
Caryopses (15 days after flowering) were harvested from einkorn line ID1331, characterised by high bread making quality (Borghi et al. 1996). Three grams of dehulled grains were frozen in liquid nitrogen, ground with mortar and pestle, and dissolved in 5 ml of 100 mM Tris–HCl, pH 8.0, 1% SDS, 100 mM LiCl, 10 mM EDTA and 200 μl 2-mercaptoethanol, in the presence of Polyclar®AT (Sigma, St Luis, Missouri). Freshly prepared phenol:chloroform:isoamyl alcohol (25:24:1, PCI) was added and the phases were mixed for 1 min, incubated for 5 min at 65°C and again mixed for 1 min. After centrifugation for 10 min at 5,000g, the phenol extraction of the supernatant was repeated five times at room temperature followed by two chloroform:isoamyl alcohol (24:1, CI) extractions. The aqueous phase was precipitated at 4°C overnight by 0.4 volumes 8 M LiCl. After centrifugation at 4°C and 19,000g for 30 min and a washing-step in 80% ethanol, the RNA-pellet was dissolved in DEPC treated water. To further reduce the amount of polysaccharides and better purify RNA, the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) was used following the protocol for “Purification of Total RNA from Plant Cells and Tissues and Filamentous Fungi” with buffer RLC.
Poly A+ RNA was then extracted using the Oligotex Kit (Qiagen, Hilden, Germany) following the “Oligotex mRNA Spin-Column Protocol” according to the manufacturer’s instructions but with an initial incubation at room temperature. To further remove traces of polysaccharides, 1/25th volume of 5 M NaCl was added and the sample was incubated for 30 min on ice, followed by one centrifugation step at 4°C and 15,000g for 10 min (Shirzadegan et al. 1991).
The purified supernatant underwent a final phenol extraction (P, PCI, 2 × CI), and the purified poly A+ RNA was precipitated by adding 2.5 volumes of ethanol at −80°C overnight. After centrifugation at 4°C and 13,000g for 20 min and a final washing step with 70% ethanol the precipitate was finally dissolved in DEPC treated water.
cDNA synthesis and sequencing
Two hundred nanograms of purified poly A+ RNA were transcribed in the first strand synthesis using the BD Clontech SMART™ cDNA synthesis kit (Clontech Laboratories, Mountain View, CA, USA). After second strand synthesis the resulting cDNA was SfiI digested, column fractionated and ligated directly into a modified pBluescript plasmid (Stratagene, La Jolla, CA, USA). The vector contains the sequence of pTriplEx2 from the EcoRI and the SalI restriction site including the SfiIA and the SfiIB restriction sites, which were cloned between the corresponding EcoRI and SalI restriction sites into the pBluescript polylinker. Single fractions of the column-separated cDNA digest were ligated and transformed in electro competent Escherichia coli dH10b cells (Invitrogen, Carlsbad, CA, USA).
About 960 randomly selected clones were sequenced, resulting in 918 expressed sequence tags (ESTs). Sequence reactions were performed using an Applied Biosystems (Weiterstadt, Germany) ABI Prism 3730xL sequencer using BigDye terminators either directly from both ends with SK and T7 primers on the plasmid DNA or with the SK primer after amplification of the insert DNA by using the T3 and T7 primer.
The sequences were processed with Applied Biosystems DNA Sequencing Analysis Software and alignments of the deduced amino acid sequences were assembled, edited and displayed using the BioEdit Sequence Alignment Editor program 7.0.9 (Hall 1999).
EST data mining
The resultant contigs were annotated according to DNA sequence homologies against the NCBI database using the BLAST method (Altschul et al. 1990, 1997).
Screening for HMW clones
Probe design
The oligonucleotide pair, AEF2 5′-CTG GGC AAC TAC AGT GTG AGC-3′ and AER3 5′-GTC TTT GTT GCT CTT GTG TTG G-3′ (Operon, Cologne, Germany) was designed to amplify a 507-bp HMW fragment (between nucleotide positions 77 and 583 of the HMW-x gene of ID1331) spanning both a 5′-conserved region and a highly repetitive region and showing the highest similarity to T. aestivum HMW glutenin subunit Ax2* gene (GenBank Number M22208). This region was amplified from three diploid wheats (T. monococcum ID1331, T. boeoticum ID 839 and T. urartu ID 1399). The amplification conditions were initial denaturation at 94°C for 3 min (denaturation 94°C 30 s/annealing 62.5°C 30 s/elongation 72°C 40 s) × 31 cycles, final elongation 72°C 6 min. All PCR products were pooled and used as probes.
Hybridization
cDNA of ID 1331 was ligated into the modified pBluescript plasmid and about 18,000 clones were spotted on Hybond™-N membranes (GE Healthcare, Chalfont St Giles, UK) in 5 × 5 double spotting design. Pooled PCR fragments (45 μl) were labelled with 50 μCi of alpha 32P dCTP using the Amersham Rediprime II Random Prime Labelling System (GE Healthcare, Chalfont St Giles, UK) following the manufacturer’s instructions. Pre-hybridisation was for 1 h at 65°C in 200 ml hybridisation solution (0.35 M sodium phosphate buffer pH 7.2, 3% SDS, 40 μg/mL salmon sperm DNA). The labelled probe was added to 300 ml hybridisation solution and incubated over night at 65°C. Four washing steps were performed: 2 × 2 × SSC, 0.1% SDS and 2 × 0.2 × SSC, 0.1% SDS in a total volume of 500 ml, at 65°C.
x-type HMW gene sequencing
PCR amplification
Specific primers were designed to amplify the complete x-type HMW gene from ID 1331. PCR primer combinations were (I) HW01 5′-ACT AAG CGG TTG GTT CTT TTT G-3′ (binding position corresponding to HMW-x for ID 1331: 04) and HW05 5′-CTG TGC CTT TGC CAC CTT TAG-3′ (2361); (II) HW08 5′-CAC CGA GCA TCA CAA ACT AGA G-3′ (−47) and HW10 5′-AAC ATG GTA TGG GCT GTC GTA G-3′ (2316); and (III) HWA 5′-AGA TGA CTA AGC GGT TGG TTC-3′ (D’Ovidio et al. 1995; −2) and HWB 5′-CTG GCT GGC CAA CAA TGC GT-3′ (D’Ovidio et al. 1995; 2427). PCR conditions were (I) 95°C for 5 min, (94°C 30 s/63°C 9 s/72°C 2 min 50 s) × 45, 72°C 7 min; (II) 95°C 5 min, (94°C 30 s/63°C 9 s/72°C 2 min 50 s) × 45, 72°C 7 min; and (III) 95°C 5 min, (94°C 30 s/63°C 9 s/72°C 2 min 50 s) × 40, 72°C 7 min. PCR reaction (20 μl) used 1 μl 10 pmol primer each, 2.5 μl 2 mM dNTPs, 2 μl 10 × Tuning Buffer (Eppendorf, Hamburg, Germany), 0.3 μl TripleMaster PCR System (Eppendorf, Hamburg, Germany), 0.9 μl 25 mM magnesium acetate (MgOAc), 0.5 μl enhancer (Invitrogen, Carlsbad, USA), 11.8 μl distilled water. PCR products were ligated into the pGEM®-T Easy Vector (Promega, Madison, USA) and transformed into E. coli dH10b cells (Invitrogen, Carlsbad, USA). Cloned PCR products were sequenced from both ends.
Exonuclease III sequencing
Nested sets of unidirectional deletions using exonuclease III in conjunction with S1 Nuclease (Henikoff 1984) were carried out on three candidate plasmids containing the HMW x-type gene. All enzymes used were from Fermentas (Hurlington, Canada). Plasmid DNAs were digested with SacI, ammonium acetate- and glycogen-precipitated and then SalI digested in Buffer O. Fragments were then separated in 1% TopVision™ LM GQ Agarose and those of 5.5 kb were excised, incubated at 70°C and Agarase digested. The DNA was re-suspended in 56 μl 1× Exonuclease III Reaction Buffer at 37°C, and 2.5 μl (493 U) Exonuclease III were added. Exonuclease III plasmid reactions of 3.0 μl were transferred at intervals of 30 s to 30 μl cold S1-Nuclease-Mix (S1-solution for 15 sample time points: 118 μl 5× S1-reaction buffer, 4 μl S1-Nuclease and 460 μl distilled water), and left on ice. The solutions were incubated for 30 min at room temperature and the reaction stopped by adding 7.2 μl S1 STOP solution (300 mM Tris, 50 mM EDTA), mixed and denaturated at 70°C for 10 min. Solutions at selected time points were ammonium acetate- and glycogen-precipitated, re-suspended in distilled water, treated with Klenow fragment for 15 min at 30°C and denaturated for 15 min at 70°C. All solutions were again precipitated and the digested plasmids re-ligated with T4 DNA ligase in 20 μl volumes overnight at 16°C. Samples (4 μl) were transformed into E. coli dH10b cells (Invitrogen, Carlsbad, USA) and DNAs extracted. The plasmids were linearized by XmnI and tested on 1% agarose gel. All plasmids containing insert lengths between 5.5 kb (t0) and 3.5 kb (empty pGEM®-T Easy Vector) were sequenced from both ends.
All sequence data have been deposited in GenBank Data library under accession FJ441077–FJ441123.
Results
cDNA library construction
A cDNA library from endosperm of T. monococcum, accession ID 1331, was produced to obtain seed storage EST sequences. The challenge was to overcome the negative effects of the large amount of polysaccharides present in the extracted material. The critical steps for success were several rounds of phenol extraction using freshly prepared PCI/CI and incubation step at 65°C. The last traces of polysaccharides were removed by adding 1/25th volume of 5 M NaCl to the sample and by incubating for 30 min on ice, followed by one centrifugation step at 4°C and 15,000g for 10 min.
Sequencing of cDNAs
Sequence reactions were performed either directly on the plasmid DNA after amplification with the pBluescript SK-primer or were based on the same SK primer after amplification of the insert DNA by using the T3 and T7 primers. In total, 960 randomly selected cDNA clones were sequenced which resulted in 918 EST sequences.
Besides storage protein cDNA sequences, clones were identified which were related to (1) carbohydrate metabolism (about 3% of sequenced clones), such as ADP-glucose-pyrophosphorylase, 6-phosphofructokinase, glyceraldehyde-3-phosphate dehydrogenase, methylmalonate semialdehyde dehydrogenase, β-amylase, granule-bound starch synthase, fructanfructosyltransferase and cellulose synthase; (2) transcription and translation mechanisms (9%) like ribosomal protein L32 and translation initiation and translation elongation factors; (3) cell metabolism (2%) like S-adenosylmethioninsynthetase, aspartate aminotransferase, O-acetylserine lyase, putative serine/threonin kinase; and (4) other cell metabolisms like betaine aldehyde dehydrogenase (BADH), late embryogenesis abundant proteins (LEA), bax inhibitor, putative kinetochore protein, hypothetic transposase, polyubiquitin, putative protein kinase C inhibitor. Twelve percent of the sequences, corresponding to 115 clones were not homologous to ESTs present in plant databases. The remaining 547 cDNA sequences were related to puroindolines (8 cases), thionins (13) and to eight genes having a low but still evident homology to true storage protein genes. The cDNA clones assigned to true storage protein genes were 518, of which 235 were either short or difficult to be assigned to either α/β-, γ-, ω-gliadins or to glutenins. The sequences assigned to α/β-, γ-, ω-gliadins and LMW glutenins were, respectively, 135, 69, 2, and 74. No full length HMW sequences were found within the 918 ESTs sequenced. The length of HMW subunits, with transcripts typically greater than 2 kb, and the repetitive nature of the sequence made full-length HMW cDNA difficult to be cloned. Full-length HMW genes were also difficult to amplify by PCR, again probably because of the length and repetitive structure between the conserved N- and C-terminal domains (Shewry et al. 2003).
Storage protein genes: gliadins and LMW glutenins
This paper addresses a simple question: how to evaluate the number of genes which encode for bona fide toxic or immunogenic gluten peptides in diploid wheat T. monococcum. This number can be approached by estimating the number of different cDNA sequences encoding for such peptides.
Based on available literature, and considering only peptides with a proved activity (either in vivo or in vitro), among α/β-gliadins of T. monococcum we identified eight antigenic peptides (numbered in italics and bold in Table 1) corresponding to the following sequences: 1: LGQ3PFP2Q2PYPQPQPF; 2: PQPQPFPSQ2PY; 3: QLQPFPQPQLPY; 4: PFRPQ2PYPQPQPQ; 5: QNPSQ3PQEQVPLVQ3; 6: QLIPCMDVVL; 7: PSGQGSFQPSQ2; 8: LGQGSFRPSQ2 N. These peptides are present in three protein molecule regions defined in this paper as “antigenic”, with the roman numbers I–III (Table 1). Region I is characterized by the combinations of peptides 1, 2, 3, 4, or 1, 2, 3, 4, 5, or 1, 2, 3, or 3, 4, 5; region II by the presence or absence of peptide 6; region III includes peptides 7 or 8. The estimate of the number of α/β-gliadin genes is 17 (Table 1). In the same Table the signal peptides, the poly glutamine regions and the C-terminal regions of the α/β-gliadin molecules are reported.
Among γ-gliadins, four antigenic peptides were found (Table 2a): peptide 9: PQ2PFPQ2PQ2; 10: Q2PQ2PFPQ; 11: PQ2SFPQ3; 12: IIQPQ2PAQ. Two antigenic regions were identified: region IV, containing the single peptides 10 or 11 or the combinations of 9 plus 10 or 10 plus 11; region V, containing peptide 12. The number of estimated γ-gliadin genes is 12. In the Table, signal peptides and the poly glutamine and C-terminal regions of γ-gliadin molecules are also reported.
Only one gene coding for ω-gliadins was present among the 237 cDNA clones sequenced. The gene encodes for the antigenic peptide 13: PQ2PFPQ2, located in antigenic region VI (Table 2b).
Among LMW glutenins, the presence of three antigenic peptides was evident (Table 3a): peptides 14: FSQ4SPF; 15: PFSQ5; 16: PFSQ4PV, which are present in the antigenic region VII alone or in the combination of peptides 14 plus 15. The estimate of the number of LMW glutenin genes is 11. Other characteristics of the seven LMW glutenin molecules are reported in Table 3a.
Storage protein genes: HMW glutenins
To uncover complete HMW sequences from ID1331 two strategies were followed: (1) HMW fragments were PCR amplified with primer combination AEF2-AER3 from three diploid wheat accessions. Two to three fragments were produced due to the binding of AER3 in different repetitive regions (for T. monococcum ID1331, fragments were of about 510, 950 and 1,700 bp. For T. boeoticum ID839, 510, 950 and 1,700 bp. For T. urartu ID1399, 550 and 1,700 bp). All fragments were pooled and used as probe to screen Hybond™-N filters for HMW clones. Forty clones were detected and eight selected. Plasmid DNA was isolated and sequenced using the SK primer. Sequencing supported the assignment of clones to y-type HMW (fragments of 372, 526, 687, 728, 819, 878, 898 and 903 bp) and a consensus sequence of 1,320 bp was obtained corresponding to positions from 658 to 1,830 of the gene encoding the HMW subunit 1Ay1 of T. urartu (gene AY245578). The 898-bp fragment had a 18-bp deletion within a 108-bp insertion unique for ID 1331 compared to other published HMW-y sequences. Thus, two different y-type sequences became available for ID1331 (Fig. S1; GenBank submission numbers FJ441119, FJ441120). The y-type sequence from T. monococcum ID 1331 has great homology with y-type sequences belonging to chromosome 1A in other Triticum species, and the closest relative to our sequences is found in T. urartu. No clone carrying an x-type HMW fragment was detected among the sequenced clones. (2) The x-type HMW gene was amplified using three PCR primer combinations and cloned. Fifty-five clones of T. monococcum line ID1331 having the expected 2.5 kb insert of the x-type HMW gene were sequenced from both ends. An exonuclease III treatment was adopted due to the existence in the gene of a highly repetitive region consisting of hexa-, nona-, and tri-peptide motifs, and a procedure of primer walking was followed. Nested sets of unidirectional deletions generated using the exonuclease III were carried out for three candidate plasmids and 89 clones were sequenced (plasmid size between 5.5 kb (t0) and 3.5 kb, empty pGEM®-T Easy Vector). All sequences were assembled in a consensus spanning positions 1–2,361 of the complete x-type HMW gene of ID1331, which has a total length of 2,430 bp (Fig. S2; GenBank submission number FJ441118). The last 69 bp were not sequenced due to the binding site of reverse PCR primers still present in the coding sequence at the 3′ end of the gene.
The sequence in Fig. S2 is the first complete x-type HMW reported for T. monococcum. The x-type gene from ID 1331 has great homology with x-type sequences belonging to chromosome 1A in other Triticum species, and the closest relative to our sequences is found in T. urartu. Relevant regions of the HMW glutenins from T. monococcum ID1331 are reported in Table 3b; one antigenic peptide was identified, peptide 17: QGYYPTSPQ, present in the antigenic region VIII (Table 3b).
Discussion
Up to date the involvement of gluten in the CD syndrome has been studied in detail in bread wheat, where a set of “toxic” and “immunogenic” peptides has been defined. The majority of the epitopes revealed “immunogenic”, i.e. able to stimulate specific T-cell lines and clones derived from jejunal mucosa or peripheral blood of celiac patients; however, only few of them were shown to be “toxic” that is able to induce mucosal damage when added in culture to duodenal mucosal biopsy or administered in vivo on proximal and distal intestine (Ciccocioppo et al. 2005).
In the present paper we listed CD antigenic epitopes found after sequencing a large number of T. monococcum cDNA clones related to seed storage proteins. As reported in Tables 1, 2 and 3, we found four bona fide toxic peptides (epitopes 1, 2, 8, 13) and 13 immunogenic peptides (epitopes 3–7, 9–17; references in the footnotes of the tables).
Information on the epitopes is poor for wheat diploid species. Mölberg et al. (2005), screening Aegilops and Triticum species related to the A, B and D genome with T-cell clones specific for 3 α- and 5 γ-gliadin epitopes, demonstrated distinct differences in the intestinal T-cell responses to the diploid species. In addition, they found that the fragments equivalent to the highly immunostimulatory 33mer peptide were absent from gluten of einkorn. Spaenij-Dekking et al. (2005) tested some accessions of 2n, 4n and 6n Triticum species by both T cell and antibody-based assays for the presence of T-cell stimulatory epitopes in gliadin and glutenins and observed a large variation in the amount of T-cell-stimulatory peptides, independent of the ploidy level. Van Herpen et al. (2006), studying alpha-gliadin genes from the A, B and D genomes of wheat, demonstrated that the set of CD epitopes was distinct for each genome.
A conclusion deriving from these studies is that einkorn, T. monococcum, has the full potential to induce the CD syndrome, as already evident for polyploid wheats (relevant references in Anderson and Wieser 2006). This is an important conclusion because recent papers (De Vincenzi et al. 1996; Pizzuti et al. 2006; Vicentini et al. 2007) claim a lack of toxicity of T. monococcum in an in vitro organ culture system, casting doubts on the capacity of patients fed with T. monococcum-derived products to develop the disease. Such a position was supported by the absence in T. monococcum of the highly immunoreactive 33-mer peptide LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF from α/β-gliadins, which was shown to be encoded by genes located on chromosome 6D (Mölberg et al. 2005). However, even after an accurate consideration of the differences between T. monococcum and other wheats, the list of epitopes in Tables 1, 2 and 3 should indicate that more experiments are needed before accepting the view that T. monococcum-derived products are less toxic or non toxic for CD patients.
We have predicted the existence of 17 α/β-, 12 γ-, 1 ω-gliadin, 11 LMW-, and 3 HMW-glutenin genes for a total of 44 genes. Some of these genes were previously located by our group in a chromosome map derived from a cross involving the same einkorn accession used in this study: in strict homology to gene loci of other Triticeae, HMW glutenin genes of T. monococcum were allocated to the long arm of chromosome 1, LMW glutenin, ω- and γ-gliadin genes to the short arm of chromosome 1 and α/β-gliadin genes to the short arm of chromosome 6 (Tänzler et al. 2002). During wheat evolution and domestication, 2n and 4n–6n wheats followed two independent paths: T. monococcum derives from the wild T. boeoticum and the domestication took place around 10000 BP (Salamini et al. 2002; Kilian et al. 2007b); domesticated 4n and 6n wheats both derive from the wild form T. dicoccoides, which in turn originated from the fusion of T. urartu and Aegilops speltoides genomes about 0.25–1.3 MYA BP (Mori et al. 1995; Huang et al. 2002; Dvorak and Akhunov 2005; Kilian et al. 2007a). Due to difference in origin, the arsenal of storage protein genes should be, to some extent, different, as stated by Wieser (2000).
Tänzler et al. (2002) showed that in einkorn the loci on the short arm of chromosome 1 greatly influence bread-making quality, measured as SDS sedimentation volume and specific sedimentation volume; they demonstrated that a large QTL for bread-making quality was consistently present across four environments on the short arm of chromosome 1, with a high probability to be represented by the LMW glutenin loci. In polyploid wheats, QTLs for bread-making quality were mapped to the long arm of chromosome 1, close to the Glu-1 locus (Sourdille et al. 1999; Perretant et al. 2000; Campbell et al. 2001; Arbelbide and Bernardo 2006). However, several studies revealed that also in polyploid wheats the allelic variation at the LMW-GS coding loci was associated with differences in dough quality (Pogna et al. 1990; Ruiz and Carrillo 1993; Gupta et al. 1994). In conclusion, we believe that, in spite of the noted differences among diploid and polyploidy wheats, the catalogue of storage protein peptides reported in this paper is sufficient to consider also T. monococcum as a source of products toxic for CD patients.
In our cDNA library we identified four i-type, one s-type and two m-type LMW glutenin sequences. Although the first i-type sequence was published in 1988 (Pitts et al. 1988), the different structure of the i-type genes, compared to m-type, did not receive enough attention: it was thought that these sequences probably belonged to truncated genes. Cloutier et al. (2001) recently reported the gene sequence and characterization of an i-type LMW-GS; expression of i-type LMW-GS genes was proven also by proteomics work (Ferrante et al. 2004) and by analysing ESTs databases (Juhász et al. 2003). Juhász and Gianibelli (2006) assumed that i-type LMW genes are characteristic of the Glu-A3 locus. Wicker et al. (2003) by sequencing a BAC contig from T. monococcum, proved the presence of three paralogous i-type genes. Our work strongly supports these findings.
The information derived from the present paper strongly suggests against the approach of breeding wheat species low in sequences noxious for CD patients by eliminating the immunogenic or toxic epitopes from their storage proteins arsenal: in fact, it seems hardly feasible to create new genotypes lacking all the 17 harmful peptides, belonging to different loci. Accordingly, the silencing, via targeted mutagenesis, of the genes giving rise to immunostimulatory sequences (Vader et al. 2003) appears also unrealistic.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Anderson RP, Wieser H (2006) Medical applications of gluten-composition knowledge. In: Wrigley C, Bekes F, Bushuk W (eds) Gliadin and glutenin. The unique balance of wheat quality. AACC International Press, St Paul, pp 387–409
Arbelbide M, Bernardo R (2006) Mixed-model QTL mapping for kernel hardness and dough strength in bread wheat. Theor Appl Genet 112:885–890
Arentz-Hansen H, Korner R, Mölberg Ø, Quarsten H, Vader W, Kooy YMC, Lundin KEA, Koning F, Roepstorff P, Sollid LM, McAdam SN (2000) The intestinal T-cell response to a α-gliadin in adult celiac disease is focused on a single deamidated glutamine targeted by tissue transglutaminase. J Exp Med 191:603–612
Arentz-Hansen H, McAdam SN, Mölberg Ø, Fleckenstein B, Lundin KEA, Jorgensen TJD, Jung G, Roepstorff P, Sollid LM (2002) Celiac lesion T cells recognize epitopes that cluster in regions of gliadins rich in proline residues. Gastroenterology 123:803–809
Borghi B, Castagna R, Corbellini M, Heun M, Salamini F (1996) Breadmaking quality of einkorn wheat (Triticum monococcum ssp. monococcum). Cereal Chem 73:208–214
Branlard G, Autran J-C, Monneveux P (1989) High molecular weight glutenin subunits in durum wheat (T. durum). Theor Appl Genet 78:353–358
Campbell KG, Finney PL, Bergman CJ, Gualberto DG, Anderson JA, Giroux MJ, Siritunga D, Zhu J, Gendre F, Roué C, Vérel A, Sorrells ME (2001) Quantitative trait loci associated with milling and baking quality in a soft × hard wheat cross. Crop Sci 41:1275–1285
Ciccocioppo R, Di Sabatino A, Corazza GR (2005) The immune recognition of gluten in coeliac disease. Clin Exp Immunol 140:408–416
Cloutier S, Ramptisch C, Penner GA, Lukow OM (2001) Cloning and expression of a LMW-i glutenin gene. J Cereal Sci 33:143–154
D’Ovidio R, Masci S (2004) The low-molecular-weight glutenin subunits of wheat gluten. J Cereal Sci 39:321–339
D’Ovidio R, Masci S, Porceddu E (1995) Development of a set of oligonucleotide primers specific for genes at the Glu-1 complex loci of wheat. Theor Appl Genet 91:189–194
De Vincenzi M, Luchetti R, Giovannini C, Pogna NE, Saponaro C, Galterio G, Gasbarrini G (1996) In vitro toxicity testing of alcohol-soluble proteins from diploid wheat Triticum monococcum in celiac disease. J Biochem Toxicol 11:313–318
Dewar DH, Amato M, Ellis HJ, Pollock EL, Gonzalez-Cinca N, Wieser H, Ciclitira PJ (2006) The toxicity of high molecular weight glutenin subunits of wheat to patients with coeliac disease. Eur J Gastroenterol Hepatol 18(5):483–491
Dvorak J, Akhunov E (2005) Tempos of gene locus delations and duplications and their relationship to recombination rate during diploid and polyploid evolution in the Aegilops–Triticum alliance. Genetics 171:323–332
Ensari A, Marsh MN, Moriarty KJ, Moore CM, Fido RJ, Tatham AS (1998) Studies in vivo of ω-gliadins in gluten sensitivity (celiac sprue disease). Clin Sci (Lond) 95:419–424
Ferrante P, Patacchini C, Masci S, D’Ovidio R, Lafiandra D (2004) LMW-i type subunits are expressed in wheat endosperm and belong to the glutenin fraction, vol 295. Royal Society of Chemistry, London, pp 136–139
Fluge O, Sletten K, Fluge G, Aksnes L, Elsayed S (1994) In vitro toxicity of purified gluten peptides tested by organ culture. J Pediatr Gastroenterol Nutr 18:186–192
Gianfrani C, Troncone R, Mugione P, Cosentini E, De Pascale M, Faruolo C, Senger S, Terrazzano G, Southwood S, Auricchi S, Sette A (2003) Celiac disease association with CD8+ T cell responses: identification of a novel gliadin-derived HLA-A2-restricted epitope. J Immunol 170:2719–2726
Gupta RB, Paul JG, Cornish GB, Palmer GA, Bekes F, Rathjen AJ (1994) Allelic variation at glutenin subunit and gliadin loci, Glu-1, Glu-3 and Gli-1 of common wheats. I. Its additive and interaction effects on dough properties. J Cereal Sci 19:9–17
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Henikoff S (1984) Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene 28:351–359
Howdle PD, Ciclitira PJ, Simpson FO (1984) Are all gliadins toxic in coeliac disese? An in vitro study of α, β, γ, ω gliadins. Scand J Gastroenterol 19:41–47
Huang S, Sirikhachornkit A, Su X, Faris J, Gill B, Haselkorn R, Gornicki P (2002) Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploidy wheat. Proc Natl Acad Sci USA 99:8133–8138
Jackson EA, Holt LM, Payne PI (1983) Characterization of high molecular weight and low molecular weight subunits of wheat endosperm by two-dimensional electrophoresis and chromosomal localization of the controlling genes. Theor Appl Genet 66:29–37
Juhász A, Gianibelli MC (2006) Low-molecular-weight glutenin subunits: insight into this abundant subunit group present in glutenin polymers. In: Wrigley CW, Békés F, Bushuk W (eds) Gliadin and glutenin: the unique balance of wheat quality. AACC International Press, St Paul, pp 171–212
Juhász A, Gardonyi M, Tamas L, Bedo Z (2003) Characterization of the promoter region of Glu-1Bx7 gene from over expressing lines of an old Hungarian wheat variety. In: Pogna NE, Romano M, Galterio G (eds) Proceedings of the 10th international wheat genet symposium, Paestum, Italy, vol 3. Istituto Sperimentale per la Cerealicoltura, Rome, pp 1348–1350
Kagnoff MF (2007) Celiac disease: pathogenesis of a model immunogenetic disease. J Clin Invest 117:41–49
Kilian B, Özkan H, Deusch O, Effgen S, Brandolini A, Kohl J, Martin W, Salamini F (2007a) Independent wheat B and G genome origins in outcrossing Aegilops progenitor haplotypes. Mol Biol Evol 24:217–227
Kilian B, Özkan H, Walther A, Kohl J, Dagan T, Salamini F, Martin W (2007b) Molecular diversity at 18 loci in 321 wild and 92 domesticated lines reveal no reduction of nucleotide diversity during Triticum monococcum (einkorn) domestication: implications for the origin of agriculture. Mol Biol Evol 24:2657–2668
Lew EJ-L, Kuzmicky DD, Kasarda DD (1992) Characterization of low molecular weight glutenin subunits by reversed-phase high-performance liquid chromatography, sodium dodecyl sulphate-polyacrylamide gel electrophoresis, and N-terminal amino acid sequencing. Cereal Chem 69:508–515
Lundin KEA, Scott H, Hansen T, Paulsen G, Halstensen TS, Fausa O, Thorsby E, Sollid LM (1993) Gliadin-specific, HLA-DQ(α1*0501, β1*0201) restricted T cells isolated from the small intestinal mucosa of celiac disease patients. J Exp Med 178:187–196
Mantzaris G, Jewell DP (1991) In-vivo toxicity of a synthetic dodecapeptide from A gliadin in patients with celiac disease. Scand J Gastroenterol 26:392–398
Masci S, Lafiandra D, Porceddu E, Lew EJ-L, Tao HP, Kasarda DD (1993) d-glutenin subunits: N-terminal sequences and evidence for the presence of cysteine. Cereal Chem 70:581–585
Masci S, Rovelli L, Kasarda DD, Vensel WH, Lafiandra D (2002) Characterization and chromosomal localisation of C-type low-molecular-weight glutenin subunits in the bread wheat cultivar Chinese Spring. Theor Appl Genet 104:422–428
Molberg Ø, Solheim Flaete N, Jensen T, Lundin KE, Arentz-Hansen H, Anderson OD, Kjersti Uhlen A, Sollid LM (2003) Intestinal T cell responses to high-molecular-weight glutenins in celiac disease. Gastroenterology 125:337–344
Mölberg Ø, Uhlen AK, Jensen T, Solheim Flaete N, Fleckenstein B, Arentz-Hansen H, Raki M, Lundin KEA, Sollid LM (2005) Mapping of gluten T cell epitopes in the bread wheat ancestors: implications for celiac disease. Gastroenterology 128:393–401
Mori N, Liu Y-G, Tsunewaki K (1995) Wheat phylogeny determined by RFLP analysis of nuclear DNA 2. Wild tetraploid wheats. Theor Appl Genet 90:129–134
Payne PI (1987) Genetics of wheat storage proteins and the effect of allelic variation on bread-making quality. Ann Rev Plant Physiol 38:141–153
Perretant MR, Cadalen T, Charmet G, Sourdille P, Nicolas P, Boeuf C, Tixier MH, Branlard G, Bernard S, Bernard M (2000) QTL analysis of bread-making quality in wheat using a doubled haploid population. Theor Appl Genet 100:1167–1175
Pitts EG, Rafalski JA, Hedgcoth C (1988) Nucleotide sequence and encoded amino acid sequence of a genomic gene region for a low molecular weight glutenin. Nucleic Acids Res 16:11376
Pizzuti D, Buda A, D’Odorico A, D’Inca R, Chiarelli S, Curioni A, Martines D (2006) Lack of intestinal mucosal toxicity of Triticum monococcum in celiac disease patients. Scand J Gastroent 41:1305–1311
Pogna NE, Autran JC, Mellini F, Lafiandra D, Feillet P (1990) Chromosome 1B-encoded gliadins and glutenin subunits in durum wheat: genetics and relationship to gluten strength. J Cereal Sci 11:15–34
Qiao S-W, Bergseng E, Molberg O, Jung G, Fleckenstein B, Sollid LM (2005) Refining the rules of gliadins T cell epitope binding to the disease-associated DQ2 molecule in celiac disease: importance of proline spacing and glutamine deamidation. J Immunol 175:254–261
Ruiz M, Carrillo JM (1993) Linkage relationships between prolamin genes on chromosomes 1A and 1B of durum wheat. Theor Appl Genet 87:353–360
Salamini F, Özkan H, Brandolini A, Schäfer-Pregl R, Martin W (2002) Genetics and geography of wild cereal domestication in the near east. Nat Genet Rev 3:429–441
Shewry PR, Halford NG (2002) Cereal seed storage proteins: structures, properties and role in grain utilization. J Exp Botany 53:947–958
Shewry PR, Halford NG, Tatham AS (1992) High molecular weight subunits of wheat glutenin. J Cereal Sci 15:105–120
Shewry PR, Halford NG, Lafiandra D (2003) Genetics of wheat gluten proteins. Adv Genet 49:111–184
Shirzadegan M, Christie P, Seemann JR (1991) An efficient method for isolation of RNA from tissue cultured plant cells. Nucleic Acids Res 19(21):6055
Sjostrom H, Lundin KE, Molberg Ø, Korner R, McAdam SN, Anthonsen D, Quarsten H, Noren O, Roepstorff P, Thorsby E, Sollid LM (1998) Identification of a gliadin T cell epitope in coeliac disease: general importance of gliadin deamidation for intestinal T cell recognition. Scand J Immunol 48:111–115
Sourdille P, Perretant MR, Charmet G, Cadalen T, Tixier MH, Joudrier P, Gautier MF, Branlard G, Bernard S, Boeuf C, Bernard M (1999) Detection of QTL for bread-making quality in wheat using molecular markers. In: Scarascia-Mugnozza GT, Porceddu E, Pagnotta MA (eds) Genetics and breeding for crop quality and resistance. Kluwer, Dordrecht, pp 361–366
Spaenij-Dekking L, Kooy-Winkelaar Y, Van Veelen P, Drijfhout JW, Jonker H, Van Soest L, Smulders MJM, Bosch D, Gilissen LJWJ, Koning F (2005) Natural variation in toxicity of wheat: potential for selection of nontoxic varieties for celiac disease patients. Gastroenterology 129:797–806
Sturgess R, Day P, Ellis HJ, Lundin KE, Gjertsen HA, Kontakou M, Ciclitira PJ (1994) Wheat peptide challenge in celiac disease. Lancet 343:758–761
Tänzler B, Esposti RF, Vaccino P, Brandolini A, Effgen S, Heun M, Schaefer-Pregl R, Borghi B, Salamini F (2002) Molecular linkage map of einkorn wheat: mapping of storage-protein and soft-glume genes and bread-making quality QTLs. Genet Res 80:131–143
Vader W, Kooy YMC, Van Veelen P, De Ru A, Harris D, Benckhuijsen W, Peña S, Mearin L, Drijfhout JW, Koning F (2002) The gluten response in children with celiac disease is directed toward multiple gliadin and glutenin peptides. Gastroenterology 122:1729–1737
Vader WL, Stepniak DT, Bunnik EM, Kooy YMC, De Haan W, Drijfhout JW, Van Veelen P, Koning F (2003) Characterization of cereal toxicity for celiac disease patients based on protein homology in grains. Gastroenterology 125:1105–1113
Van de Wal Y, Kooy YMC, van Veelen PA, Peña SA, Mearin LM, Molberg Ø, Lundin KEA, Sollid LM, Mutis T, Benckhuijsen WE, Drijfhout JW, Koning F (1998) Small intestinal T cells of celiac disease patients recognize a natural pepsin fragment of gliadin. Proc Natl Acad Sci USA 95:10050–10054
Van de Wal Y, Kooy YMC, van Veelen PA, Vader W, August SA, Drijfhout JW, Peña SA, Koning F (1999) Glutenin is involved in the gluten-driven mucosal T cell response. Eur J Immunol 29:3133–3139
van Herpen TWJM, Goryunova SV, van der Schoot J, Mitreva M, Salentijn E, Vorst O, Schenk MF, van Veelen PA, Koning F, van Soest LJM, Vosman B, Bosch D, Hamer RJ, Gilissen LJWJ, Smulders MJM (2006) Alpha-gliadin genes from the A, B, and D genomes of wheat contain different sets of celiac disease epitopes. BMC Genomics 7:1–13
Vicentini O, Maialetti F, Gazza L, Silano M, Dessi M, De Vincenti M, Pogna NE (2007) Environmental factors of celiac disease: cytotoxicity of hulled wheat species Triticum monococcum, T. turgidum ssp. dicoccum and T. aestivum ssp. spelta. J Gastroent Hepat 22:1816–1822
Waines JG, Payne PI (1987) Electrophoretic analysis of the high molecular weight glutenin subunits of T. monococcum, T. urartu, and the A genome of bread wheat (T. aestivum). Theor Appl Genet 74:71–76
Wicker T, Yahiaoui N, Guyot R, Schlagenhauf E, Liu Z-D, Dubcovsky J, Keller B (2003) Rapid genome divergence at orthologous low molecular weight loci of the A and Am genomes of wheat. Plant Cell 15:1186–1197
Wieser H (2000) Comparative investigations of gluten protein from different wheat species. I. Qualitative and quantitative composition of gluten protein types. Eur Food Res Technol 211:262–268
Wieser H (2004) Celiac disease. In: Wrigley C, Corke H, Walker CE (eds) Encyclopedia of grain science. Elsevier, Amsterdam, pp 179–187
Wieser H, Koehler P (2008) The biochemical basis of celiac disease. Cereal Chem 85(1):1–13
Wrigley CW, Bekes F, Bushuk W (2006) Gluten: a balance of gliadin and glutenin. In: Wrigley C, Bekes F, Bushuk W (eds) Gliadin and glutenin. The unique balance of wheat quality. AACC International Press, St Paul, pp 3–32
Acknowledgments
We thank Sigi Effgen, Jutta Schütze and Isabell Fuchs for excellent technical assistance. The work was supported by grants from the Deutsche Forschungsgemeinschaft (SSP-1127).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
438_2008_412_MOESM1_ESM.rtf
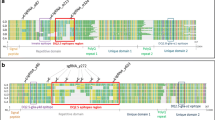
Figure S1. Multiple sequence alignment of y-type HMW glutenin genes encoded by the A genome of different Triticum species (AES: T. aestivum ssp. aestivum; SPE: T. aestivum ssp. spelta; TUR: T. turgidum; DIC: T. turgidum ssp. dicoccum; URA: T. urartu; MON: T. monococcum ssp. monococcum). Only full-length clones present in literature are reported. Clones FJ441119 and FJ441120 refer to our sequences, as submitted to GenBank. (RTF 2.21 MB)
438_2008_412_MOESM2_ESM.rtf
Figure S2. Multiple sequence alignment of x-type HMW glutenin genes encoded by the A genome of different Triticum species (AES: T. aestivum ssp. aestivum; TUR: T. turgidum; TIM: T. timopheevii; BOE: T. monococcum ssp boeoticum; URA: T. urartu; MON: T. monococcum ssp. monococcum). Only full-length clones present in literature are reported. Clone FJ441118 refer to our sequence, as submitted to GenBank. (RTF 991 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Vaccino, P., Becker, HA., Brandolini, A. et al. A catalogue of Triticum monococcum genes encoding toxic and immunogenic peptides for celiac disease patients. Mol Genet Genomics 281, 289–300 (2009). https://doi.org/10.1007/s00438-008-0412-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-008-0412-8