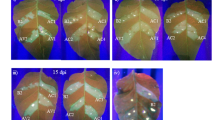
Abstract
Key message
Expression of artificial microRNA targeting ATP binding domain of AC1 in transgenic tomato confers resistance to Tomato leaf curl disease without impacting the yield of tomato.
Abstract
Tomato curl leaf disease caused by Tomato leaf curl virus (ToLCV) is a key constraint to tomato cultivation worldwide. Engineering transgenic plants expressing artificial microRNAs (amiRNAs) against the AC1 gene of Tomato leaf curl New Delhi virus (ToLCNDV), which is important for virus replication and pathogenicity, would consequently confer virus resistance and reduce crop loss in the economically important crops. This study relates to an amiRNA developed on the sequence of Arabidopsis miRNA319a, targeting the ATP/GTP binding domain of AC1 gene of ToLCNDV. The AC1-amiR was found to regulate the abundance of AC1, providing an excellent strategy in providing defense against ToLCNDV. Transgenic lines over-expressing AC1-amiR, when challenged with ToLCNDV, showed reduced disease symptoms and high percentage resistance ranging between ∼ 40 and 80%. The yield of transgenic plants was significantly higher upon ToLCNDV infection as compared to the non-transgenic plants. Although the natural resistance resources against ToLCNDV are not available, this work streamlines a novel amiRNA-based mechanism that may have the potential to develop viral resistance strategies in tomato, apart from its normal symptom development properties as it is targeting the conserved region against which higher accumulation of small interfering RNAs (siRNA) occurred in a naturally tolerant tomato cultivar.










Similar content being viewed by others
References
Ai T, Zhang L, Gao Z, Zhu CX, Guo X (2011) Highly efficient virus resistance mediated by artificial microRNAs that target the suppressor of PVX and PVY in plants. Plant Biol 13:304–316
Amari K, Gonzalez-Ibeas D, Gomez P et al (2008) Tomato torrado virus is transmitted by Bemisia tabaci and infects pepper and eggplant in addition to tomato. Plant Dis 92:1139
Ali I, Amin I, Briddon RW, Mansoor S (2013) Artificial microRNA-mediated resistance against the monopartite begomovirus cotton leaf curl Burewala virus. Virol J 10:231
Burland TG (2000) DNASTAR’s laser gene sequence analysis software. Methods Mol Biol 132:71–91
Chakraborty S (2008) Tomato leaf curl viruses from India (Geminiviridae). In: Mahy BWJ, Van Regenmortel MHV (eds) Encyclopedia of virology. Elsevier, Oxford, pp 124–133
Chan SWL, Henderson IR, Jacobsen SE (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev 6:351–360
Dasgupta A, Sinha SK, Praveen S (2004) Structure of replication initiator protein unites diverse viruses causing tomato leaf curl disease (ToLCD). Plant Sci 166:1063–1067
de Ronde D, Butterbach P, Kormelink R (2014) Dominant resistance against plant viruses. Front Plant Sci 27(5):307
Deveson I, Li JY, Millar AA (2013) MicroRNAs with analogous target complementarities perform with highly variable efficacies in Arabidopsis. FEBS Lett 587:3703–3708
Di Nicola E, Tavazza M, Lucioli A, Salandri L, Ilardi V (2014) Robust RNA silencing-mediated resistance to Plum pox virus under variable abiotic and biotic conditions. Mol Plant Pathol 15:841–847
Ding SW, Voinnet O (2007) Antiviral immunity directed by small RNAs. Cell 130:413–426
Duan CG, Wang CH, Fang RX, Guo HS (2008) Artificial microRNAs highly accessible to targets confer efficient virus resistance in plants. J Virol 82:11084–11095
Fahim M, Millar AA, Wood CC, Larkin PJ (2012) Resistance to Wheat streak mosaic virus generated by expression of an artificial polycistronic microRNA in wheat. Plant Biotechnol J 10:150–163
Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S (2013) Geminiviruses: masters at redirecting and reprogramming plant processes. Nat Rev Microbiol 11:777–788
Hu Q, Niu Y, Zhang K, Liu Y, Zhou X (2011) Virus-derived transgenes expressing hairpin RNA give immunity to Tobacco mosaic virus and Cucumber mosaic virus. Virol J 8:41
Jelly NS, Schellenbaum P, Walter B, Maillot P (2012) Transient expression of artificial microRNAs targeting Grapevine fanleaf virus and evidence for RNA silencing in grapevine somatic embryos. Transgenic Res 21:1319–1327
Kis A, Tholt G, Ivanics M, Varallyay E, Jenes B, Havelda Z (2016) Polycistronic artificial miRNA-mediated resistance to Wheat dwarf virus in barley is highly efficient at low temperature. Mol Plant Pathol 17:427–437
Kourelis J, van der Hoorn RAL (2018) Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30:285–299
Li JF, Chung HS, Niu YJ, Bush J, McCormack M, Sheen J (2013) Comprehensive protein-based artificial microRNA screens for effective gene silencing in plants. Plant Cell 25:1507–1522
Li JF, Zhang DD, Sheen J (2014) Epitope-tagged protein-based artificial miRNA screens for optimized gene silencing in plants. Nat Protoc 9:939–949
Liu L, Gu Q, Ijaz R, Zhang J, Ye Z (2016) Generation of transgenic watermelon resistance to Cucumber mosaic virus facilitated by an effective Agrobacterium-mediated transformation method. Sci Hortic 205:32–38
Lu C, Meyers BC, Green PJ (2007) Construction of small RNA cDNA libraries for deep sequencing. Methods 43:110–117
Matzke MA, Kanno T, Matzke AJ (2015) RNA-Directed DNA Methylation: the evolution of a complex epigenetic pathway in flowering plants. Annu Rev Plant Biol 66:243–267
Mehta D, Stürchler A, Anjanappa RB, Zaidi SS, Hirsch-Hoffmann M, Gruissem W, Vanderschuren H (2019) Linking CRISPR-Cas9 interference in cassava to the evolution of editing-resistant geminiviruses. Genome Biol 20:80
Meyers BC, Axtell MJ (2019) MicroRNAs in plants: key findings from the early years. Plant Cell 31:1206–1207
Mitter N, Zhai Y, Bai AX, Chua K, Eid S, Constantin M, Mitchell R, Pappu HR (2016) Evaluation and identification of candidate genes for artificial microRNA mediated resistance to tomato spotted wilt virus. Virus Res 211:151–158
Moriones E, Navas-Castillo J (2000) Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Res 71:123–134
Moriones E, Praveen S, Chakraborty S (2017) Tomato leaf curl New Delhi virus: an emerging virus complex threatening vegetable and fiber crops. Viruses 9:264
Niu QW, Lin SS, Reyes JL, Chen KC, Wu HW, Yeh SD, Chua NH (2006) Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat Biotechnol 24:1420–1428
Ossowski S, Schwab R, Weigel D (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53:674–690
Panno S, Iacono G, Davino M, Marchione S, Zappardo V, Bella P, Tomassoli L, Accotto GP, Davino S (2016) First report of Tomato leaf curl New Delhi virus affecting zucchini squash in an important horticultural area of southern Italy. New Dis Rep 33:6
Petchthai U, Yee CSL, Wong SM (2018) Resistance to CymMV and ORSV in artificial microRNA transgenic Nicotiana benthamiana plants. Sci Rep 8:9958
Porebski S, Bailey LG, Baurn BR (1997) Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep 15:8–15
Prasad A, Sharma N, Hari-Gowthem G, Muthamilarasan M, Prasad M (2020) Tomato yellow leaf curl virus: impact, challenges, and management. Trends Plant Sci S1360–1385(20):30115–30121
Qu J, Ye J, Fang R (2007) Artificial microRNA-mediated virus resistance in plants. J Virol 81:6690–6699
Rodríguez-Negrete EA, Lozano-Duran R, Piedra-Aguilera A, Cruzado L, Bejarano ER, Castillo AG (2013) Geminivirus Rep protein interferes with the plant DNA methylation machinery and suppresses transcriptional gene Silencing. New Phytol 199:464–475
Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25:2383–2399
Ruhel R, Chakraborty S (2019) Multifunctional roles of geminivirus encoded replication initiator protein. Virus Dis 30:66–73
Sahu PP, Sharma N, Puranik S, Prasad M (2014) Post-transcriptional and epigenetic arms of RNA silencing: a defense machinery of naturally tolerant tomato plant against Tomato leaf curl New Delhi virus. Plant Mol Biol Rep 32:1015–1029
Sahu PP, Rai NK, Chakraborty S (2010) Tomato cultivar tolerant to Tomato leaf curl New Delhi virus infection induces virus-specific short interfering RNA accumulation and defence-associated host gene expression. Mol Plant Pathol 11(4):531–544
Schmittgen T, Livak K (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108
Sharma N, Prasad M (2017) An insight into plant–Tomato leaf curl New Delhi virus interaction. The Nucleus 60:335–348
Sharma MK, Solanke AU, Jani D, Singh Y, Sharma AK (2009) A simple and efficient Agrobacterium-mediated procedure for transformation of tomato. J Biosci 34:423–433
Sharma N, Sahu PP, Puranik S, Prasad M (2013) Recent advances in plant-virus interaction with emphasis on small interfering RNAs (siRNAs). Mol Biotechnol 55:63–77
Sorab SS, Karim S, Varma A, Azhar EI, Mandal B, Abuzenadah AM, Chaudhary AG (2013) Factors affecting sap transmission of Tomato leaf curl New Delhi begomovirus infecting sponge gourd in India. Phytoparasitica 41:591–592
Southern E (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 98:503–517
Sun L, Lin C, Du J, Song Y, Jiang M, Liu H, Zhou S (2016) Dimeric artificial microRNAs mediate high resistance to RSV and RBSDV in transgenic rice plants. Plant Cell Tissue Organ Cult 126:127–139
Tiwari AK, Sharma PK, Khan MS, Snehi SK, Raj SK, Rao GP (2010) Molecular detection and identification of Tomato leaf curl New Delhi virus isolate causing yellow mosaic disease in bitter gourd (Momordica charantia), a medicinally important plant in India. Med Plants 2:117–123
Vu TV, Roy Choudhury N, Mukherjee SK (2013) Transgenic tomato plants expressing artificial microRNAs for silencing the pre-coat and coat proteins of a begomovirus, Tomato leaf curl New Delhi virus, show tolerance to virus infection. Virus Res 172:35–45
Wagaba H, Patil BL, Mukasa S, Alicai T, Fauquet CM, Taylor NJ (2016) Artificial microRNA-derived resistance to Cassava brown streak disease. J Virol Methods 231:38–43
Wong G, Alonso-Peral M, Bingjun L, Junyan L, Millar AA (2018) MicroRNA MIMIC binding sites: minor flanking nucleotide alterations can strongly impact MIMIC silencing efficacy in Arabidopsis. Plant Direct 2:e00088
Xu P, Zhang Y, Kang L, Roossinck MJ, Mysore KS (2006) Computational estimation and experimental verification of off-target silencing during posttranscriptional gene silencing in plants. Plant Physiol 142:429–440
Zhang N, Zhang D, Chen SL, Gong BQ, Guo Y, Xu L, Zhang XN, Li JF (2018) Engineering artificial microRNAs for multiplex gene silencing and simplified transgenic screen. Plant Physiol 178:989–1001
Zvereva AS, Pooggin MM (2012) Silencing and innate immunity in plant defense against viral and non-viral pathogens. Viruses 4:2578–2597
Acknowledgements
The authors’ work in the area of plant–virus interaction is supported by the JC Bose Fellowship (JCB/2018/00000l) from Science and Engineering Research Board (SERB), Govt. of India, India and core grant of National Institute of Plant Genome Research (NIPGR), New Delhi. The authors thank Dr. Muthamilarasan Mehanathan, School of Life Sciences, University of Hyderabad, India, for critically reading the manuscript. The authors are also thankful to DBT-eLibrary Consortium (DeLCON) for providing access to e-resources.
Author information
Authors and Affiliations
Contributions
MP and NS conceived and designed the experiments; NS conducted the experiments and analyzed data; NS and MP wrote the manuscript. All the authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Communicated by Neal Stewart.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
299_2020_2584_MOESM3_ESM.tif
Supplementary file3 Supplementary Fig. S1. (A) Schematic representation of AC1 protein showing conserved motifs I–III required for dsDNA binding and the ATP-binding domain. (B) Multiple sequence alignment of amino acid (AA) sequence and (C) nucleotide sequence of AC1 from ToLCV strains causing the tomato leaf curl disease ToLCNDV-Mild (ToLCNDV-Mld; U15016), ToLCNDV-Lucknow (ToLCNDV-Luc; Y16421), ToLCNDV-Karnataka (ToLCNDV_Kar; NC003897), ToLCNDV-Jabalpur (ToLCNDV-Jb; AY260504), ToLCNDV- IARI (ToLCNDV-IARI; AF524893), ToLCNDV-Gujarat (ToLCNDV-Guj; AAM21569), ToLCNDV-Dharwad (ToLCNDV-Dh; AY260505), ToLCNDV-Bangladesh1 (ToLCNDV_Ban1; Z48182), ToLCNDV-Bangladesh (ToLCNDV_Ban; EF450316), ToLCBV-Bangladesh4 (ToLCBV-Ban4; AF165098), ToLCNDV-Bangladesh5 (ToLCBV-Ban5; AF295401), ToLCNDV-Israel (P27259). (D) Multiple sequence alignment of amino acid (AA) sequence and (E) nucleotide sequence of AC1 from different begomoviruses including Tomato Yellow Leaf Curl Virus-Almeria (TYLCV_Almeria; AJ489258), Tomato Yellow Leaf Curl Virus-Almeria (TYLCV; FJ956702), Tomato Golden Mosaic Virus (TGMV; NC001507), Squash leaf curl China virus (SLCCNV; KC222956) Tomato yellow leaf curl China virus (TYLCCNV; CAC85509), Tomato yellow leaf curl Sardinia virus (TYLSCV; AAA47955), African cassava mosaic virus (ACMV; AAD40938), Cotton leaf curl virus (CtLCV; KC412251), Pepper huasteco yellow vein virus (PHYVV; AAL02410), and Tomato mottle virus (ToMoV; AAC32414). The AA and nucleotide residues highlighted in black are non-conserved. The AA and nucleotide residues within the red box are the target site for AC1-amiR1 and within the blue box is the target site for AC1-amiR2. (TIF 15694 kb)
299_2020_2584_MOESM4_ESM.tif
Supplementary file4 Supplementary Fig. S2. Stem-loop structure of precursor ath-miR219, AC1-amiR1 and AC1-amiR2 predicted by Mfold software. The highlighted part indicates the mature sequence of ath-miRNA319 and amiRNAs. (TIF 1403 kb)
299_2020_2584_MOESM5_ESM.tif
Supplementary file5 Supplementary Fig. S3. PCR amplification profile using vector specific primers of kanamycin gene (448bp) from genomic DNA of transgenic lines (L1-L7), Plasmid control (PC) and non-transformed control (NTC) plants. (TIF 695 kb)
299_2020_2584_MOESM6_ESM.tif
Supplementary file6 Supplementary Fig. S4. Southern blot analysis for copy number of transgene in transgenic lines. Genomic DNA was isolated from leaves of non-transformed control (NTC) and transgenic plants (L1-L6) plants, and digested with EcoRI, which cut T-DNA at single site. Digested genomic DNA was separated on 0.8%agarose gel. (A) Blot developed by using 448 bp kanamycin amplicon (NPT-II) for probing the digested genomic DNA. (B) The EtBR stained agarose gel depicting equal loading of digested DNA from different tomato samples. (TIF 1993 kb)
299_2020_2584_MOESM7_ESM.tif
Supplementary file7 Supplementary Fig. S5. Expression analysis of AC1-amiR in transgenic lines (T2:L1- L6) and non-transformed control plants by Northern blot. Antisense of mature sequence of AC1-amiR1 was used as probe. EtBr-stained rRNA was used as loading control. (TIF 255 kb)
299_2020_2584_MOESM8_ESM.tif
Supplementary file8 Supplementary Fig. S6. Southern blot analysis for virus accumulation (A) the total DNA of transgenic (T2: L1_T, L4_T and L6_T), non-transformed control (NTC_T) and cultivar H-88-78-1 (H_T) tomato plants after 21dpi digested with EcoRV. (B) Undigested total DNA of transgenic (T1: L1_T, L4_T and L6_T), non-transformed control (NTC_T) and cultivar H-88-78-1 (H_T) tomato plants after 21dpi. Different forms of ToLCNDV genome is shown as open circular (OC), linear (Lin), supercoiled (SC) and single strand (SS). Equivalent loading of DNA was shown in the Ethidium bromide stained DNA. (TIF 2653 kb)
299_2020_2584_MOESM9_ESM.tif
Supplementary file9 Supplementary Fig. S7. Expression analysis of AC1 in virus infected transgenic lines (L1, L4 and L6), H-88-78-1 and non-transformed control plants after 21dpi (A) T2 and (B) T1. EtBr-stained rRNA is shown as the equivalent loading control for the experiment. (TIF 731 kb)
Rights and permissions
About this article
Cite this article
Sharma, N., Prasad, M. Silencing AC1 of Tomato leaf curl virus using artificial microRNA confers resistance to leaf curl disease in transgenic tomato. Plant Cell Rep 39, 1565–1579 (2020). https://doi.org/10.1007/s00299-020-02584-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-020-02584-2