Abstract
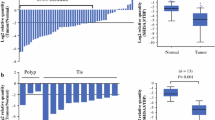
The biological mechanisms underlying resistance to tamoxifen are of considerable clinical significance. However, little is known about the correlation between tamoxifen resistance and methylation of genes related to drug-metabolizing enzymes. To address this issue, we examined the methylation pattern and expression of the selected genes coding for drug-metabolizing enzymes, including COMT, CYP1A1, CYP2D6, NAT1, and SULT1A1 in tamoxifen-resistant and control breast cancers. Bisulfite genomic sequencing and methylation-specific PCR were carried out to evaluate the methylation patterns of the five genes from control (n = 74) and tamoxifen-resistant tissues (n = 37) chosen by an age-matched sampling method. Also, end-point reverse transcriptase polymerase chain reaction (RT-PCR) and real-time RT-PCR were performed to determine RNA expression of the genes. Bisulfite genomic sequencing revealed methylation of the NAT1 gene in 25 of the control cancers (33.8%) and 23 of the resistant tumors (62.2%). Of the five genes, only NAT1 showed a significant lower methylation rate in the control group than in the resistant group (p = 0.004). No significant difference of the methylation rate was found in the other four genes including COMT, CYP1A1, CYP2D6, and SULT1A1 (p > 0.05). Furthermore, the expression rate of NAT1 mRNA was lower in the tumors from the resistant group than in control tumors (28.6% vs. 65.2%, p = 0.031). Real-time RT-PCR analysis demonstrated that the NAT1 gene was more down-regulated in resistant tissues than in control group (p = 0.023). Moreover, malignant cells from the resistant cases demonstrated a higher percentage of positive staining for Ki67 (p = 0.001) and cyclin D1 (p = 0.043) than those from the control group. Taken together, the higher methylation rate of the NAT1 gene is related to tamoxifen resistance, and this fact supports the hypothesis that hypermethylation of the NAT1 gene might affect the initiation of tamoxifen resistance.





Similar content being viewed by others
Abbreviations
- CpG:
-
Cytosine guanine dinucleotide
- ER:
-
Estrogen receptor
- MSP:
-
Methylation-specific PCR
- PgR:
-
Progesterone receptor
References
Lohrisch C, Piccart MJ (2001) Standard medical treatment for early breast cancer. Eur J Cancer 37(Suppl 7):S45–S58
MacGregor JI, Jordan VC (1998) Basic guide to the mechanisms of antiestrogen action. Pharmacol Rev 50:151–196
Osborne CK, Coronado E, Allred DC, Wiebe V, DeGregorio M (1991) Acquired tamoxifen resistance: correlation with reduced breast tumor levels of tamoxifen and isomerization of trans-4-hydroxytamoxifen. J Natl Cancer Inst 83:1477–1482
Takimoto GS, Graham JD, Jackson TA, Tung L, Powell RL, Horwitz LD, Horwitz KB (1999) Tamoxifen resistant breast cancer: coregulators determine the direction of transcription by antagonist-occupied steroid receptors. J Steroid Biochem Mol Biol 69:45–50
Kato S, Endoh H, Masuhiro Y, Kitamoto T, Uchiyama S, Sasaki H, Masushige S, Gotoh Y, Nishida E, Kawashima H, Metzger D, Chambon P (1995) Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science 270:1491–1494
Cariou S, Donovan JC, Flanagan WM, Milic A, Bhattacharya N, Slingerland JM (2000) Down-regulation of p21WAF1/CIP1 or p27Kip1 abrogates antiestrogen-mediated cell cycle arrest in human breast cancer cells. Proc Natl Acad Sci USA 97:9042–9046
Ali S, Coombes RC (2002) Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer 2:101–112
Williams JA, Phillips DH (2000) Mammary expression of xenobiotic metabolizing enzymes and their potential role in breast cancer. Cancer Res 60:4667–4677
Osborne CK, Wiebe VJ, McGuire WL, Ciocca DR, DeGregorio MW (1992) Tamoxifen and the isomers of 4-hydroxytamoxifen in tamoxifen-resistant tumors from breast cancer patients. J Clin Oncol 10:304–310
Feinberg AP, Tycko B (2004) The history of cancer epigenetics. Nat Rev Cancer 4:143–153
Esteller M (2007) Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet 8:286–298
Herman JG, Baylin SB (2003) Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 349:2042–2054
Kang JH, Kim SJ, Noh DY, Park IA, Choe KJ, Yoo OJ, Kang HS (2001) Methylation in the p53 promoter is a supplementary route to breast carcinogenesis: correlation between CpG methylation in the p53 promoter and the mutation of the p53 gene in the progression from ductal carcinoma in situ to invasive ductal carcinoma. Lab Invest 81:573–579
Kron K, Pethe V, Briollais L, Sadikovic B, Ozcelik H, Sunderji A, Venkateswaran V, Pinthus J, Fleshner N, van der Kwast T, Bapat B (2009) Discovery of novel hypermethylated genes in prostate cancer using genomic CpG island microarrays. PLoS ONE 4:e4830
Ji JW, Yang HL, Kim SJ (2006) Analysis of cag-8: a novel poly(Q)-encoding gene in the mouse brain. Biochem Biophys Res Commun 346:1254–1260
Jacobs TW, Gown AM, Yaziji H, Barnes MJ, Schnitt SJ (1999) Specificity of HercepTest in determining HER-2/neu status of breast cancers using the United States Food and Drug Administration-approved scoring system. J Clin Oncol 17:1983–1987
Dupret JM, Goodfellow GH, Janezic SA, Grant DM (1994) Structure-function studies of human arylamine N-acetyltransferases NAT1 and NAT2. Functional analysis of recombinant NAT1/NAT2 chimeras expressed in Escherichia coli. J Biol Chem 269:26830–26835
Butcher NJ, Boukouvala S, Sim E, Minchin RF (2002) Pharmacogenetics of the arylamine N-acetyltransferases. Pharmacogenomics J 2:30–42
Minchin RF (1995) Acetylation of p-aminobenzoylglutamate, a folic acid catabolite, by recombinant human arylamine N-acetyltransferase and U937 cells. Biochem J 307(Pt 1):1–3
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D (2000) Molecular portraits of human breast tumours. Nature 406:747–752
Dolled-Filhart M, Ryden L, Cregger M, Jirstrom K, Harigopal M, Camp RL, Rimm DL (2006) Classification of breast cancer using genetic algorithms and tissue microarrays. Clin Cancer Res 12:6459–6468
Bieche I, Girault I, Urbain E, Tozlu S, Lidereau R (2004) Relationship between intratumoral expression of genes coding for xenobiotic-metabolizing enzymes and benefit from adjuvant tamoxifen in estrogen receptor alpha-positive postmenopausal breast carcinoma. Breast Cancer Res 6:R252–R263
Goetz MP, Knox SK, Suman VJ, Rae JM, Safgren SL, Ames MM, Visscher DW, Reynolds C, Couch FJ, Lingle WL, Weinshilboum RM, Fritcher EG, Nibbe AM, Desta Z, Nguyen A, Flockhart DA, Perez EA, Ingle JN (2007) The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat 101:113–121. doi:10.1007/s10549-006-9428-0
Nowell S, Sweeney C, Winters M, Stone A, Lang NP, Hutchins LF, Kadlubar FF, Ambrosone CB (2002) Association between sulfotransferase 1A1 genotype and survival of breast cancer patients receiving tamoxifen therapy. J Natl Cancer Inst 94:1635–1640
Brockdorff BL, Skouv J, Reiter BE, Lykkesfeldt AE (2000) Increased expression of cytochrome p450 1A1 and 1B1 genes in anti-estrogen-resistant human breast cancer cell lines. Int J Cancer 88:902–906
Adam PJ, Berry J, Loader JA, Tyson KL, Craggs G, Smith P, De Belin J, Steers G, Pezzella F, Sachsenmeir KF, Stamps AC, Herath A, Sim E, O'Hare MJ, Harris AL, Terrett JA (2003) Arylamine N-acetyltransferase-1 is highly expressed in breast cancers and conveys enhanced growth and resistance to etoposide in vitro. Mol Cancer Res 1:826–835
Hui R, Finney GL, Carroll JS, Lee CS, Musgrove EA, Sutherland RL (2002) Constitutive overexpression of cyclin D1 but not cyclin E confers acute resistance to antiestrogens in T-47D breast cancer cells. Cancer Res 62:6916–6923
Ishii Y, Waxman S, Germain D (2008) Tamoxifen stimulates the growth of cyclin D1-overexpressing breast cancer cells by promoting the activation of signal transducer and activator of transcription 3. Cancer Res 68:852–860
Acknowledgement
These studies were supported by a grant from the National Cancer Center, Korea.
Disclosure of potential conflict of interests
The authors declare no conflict of interests related to this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
This work is supported by the National Cancer Center Grant.
Rights and permissions
About this article
Cite this article
Kim, S.J., Kang, HS., Jung, SY. et al. Methylation patterns of genes coding for drug-metabolizing enzymes in tamoxifen-resistant breast cancer tissues. J Mol Med 88, 1123–1131 (2010). https://doi.org/10.1007/s00109-010-0652-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-010-0652-z