Abstract
The interaction of electromagnetic (EM) fields with the human body during magnetic resonance imaging (MRI) is complex and subject specific. MRI radiofrequency (RF) coil performance and safety assessment typically includes numerical EM simulations with a set of human body models. The dimensions of mesh elements used for discretization of the EM simulation domain must be adequate for correct representation of the MRI coil elements, different types of human tissue, and wires and electrodes of additional devices. Examples of such devices include those used during electroencephalography, transcranial magnetic stimulation, and transcranial direct current stimulation, which record complementary information or manipulate brain states during MRI measurement. The electrical contact within and between tissues, as well as between an electrode and the skin, must also be preserved. These requirements can be fulfilled with anatomically correct surface-based human models and EM solvers based on unstructured meshes. Here, we report (i) our workflow used to generate the surface meshes of a head and torso model from the segmented AustinMan dataset, (ii) head and torso model mesh optimization for three-dimensional EM simulation in ANSYS HFSS, and (iii) several case studies of MRI RF coil performance and safety assessment.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
The interaction of radio frequency (RF) electromagnetic (EM) fields with the human body during magnetic resonance imaging (MRI) is complex and subject specific. The specific absorption rate (SAR) used as the safety limit in MRI is also subject specific, especially at RF above 100 MHz [1]. Safety limits based on the SAR in MRI are typically derived from three-dimensional (3D) numerical EM simulations of MRI RF transmit coils loaded with human body models [2,3,4,5,6,7].
An increasing number of MRI investigations that study the human brain employ multimodal setups, where additional devices are used to record complementary information or manipulate brain states [8,9,10], examples include electroencephalography (EEG), transcranial magnetic stimulation (TMS), and transcranial direct current stimulation (tDCS). This requires a dedicated setup of wires and electrodes that are in contact with human skin. For example, a tDCS setup includes two external wires and electrodes. The wires enter the MRI RF transmitter coil’s effective exposure volume and operate as an antenna, the performance of which depends on the relative positioning of the wires and the human body, patient landmark position, and the quality of the electrical contact between the electrode and skin. Assessing the RF safety of a device that is in electrical contact with the skin during an MRI examination requires the evaluation of RF-induced heating of human tissue located near the contact area.
An increasing number of MRI examinations are being performed on patients with an active implantable medical device (AIMD) , for example, a cardiac pacemaker or deep brain stimulator, or a passive implant, such as an orthopedic hip implant [11,12,13]. One of the major safety concerns for ensuring safe examinations of such patients is the evaluation of in vivo RF-induced heating of tissue near the lead electrode, which can result in tissue damage.
Due to the complexity of assessing MRI RF-induced heating in vivo, 3D EM and transient thermal co-simulation is used to assess RF-induced heating of implanted devices and devices that bring electrodes into contact with human skin [14, 15].
When modeling RF-induced heating during MRI, a computational EM solver is used to compute the absorption of EM energy in different types of human tissue. The volume and surface losses from 3D EM simulations act as thermal sources in tissue heating calculations. Volume losses in human tissue substantially depend on tissue geometries and electrical properties. For example, (i) in a patient undergoing an MRI at a head landmark position, the cerebrospinal fluid (CSF) space must be a continuous medium in the numerical domain to excite a significant current; (ii) electrical properties of the skin and underlying tissues, especially fat, differ significantly and volume losses depend on tissue geometries, (iii) the correct skin thickness is very important when assessing MRI RF safety for devices where electrodes are in contact with human skin.
Different numerical approaches can be applied to perform 3D EM and transient thermal co-simulation for simple geometrical objects, for example, a phantom as defined in ASTM F2182a-11 [16]. However, reliable simulations of realistic human models require a correct match between solver capabilities and geometrical properties of the human models under investigation.
To accurately represent individual tissue structures in a patient-specific human model, they must first be segmented from imaging data. Most imaging data are voxel-based data obtained, for example, from MRI scans, high-resolution cryosection image datasets, or histological sections. Therefore, most available numerical human models are voxel-based geometries [17].
Voxel-based human models are commonly simulated using time-domain solvers, in most cases these are finite-difference time-domain (FDTD) or finite integration technique (FIT) solvers, and use hexahedral meshes. The hexahedral mesh results in a staircased discretization of the surfaces of curved structures.
The size of the hexahedral mesh elements must be substantially smaller than the thickness of the coil’s conductive elements, the thickness of relevant thin human tissue (e.g., CSF and skin), the wire diameters, and electrode thickness of EEG or tDCS setups to maintain the precision of the geometric model. Structures that are thinner than the employed resolution are undersampled in the mesh and, therefore, appear as being separated, noncontinuous segments within a space. In this case, correct electrical and thermal contact between anatomically connected tissue parts or between an electrode and human tissue are not ensured.
In most common implementations of time-domain solvers, the size of the hexahedral mesh elements must be uniform for a given mesh line. Thus, using small-sized hexahedral mesh elements for some objects results in meshing practically the entire numerical domain with small mesh elements. Because the simulation time of a time-domain solver is proportional to the number of mesh elements and is inversely proportional to the smallest-sized mesh elements, correct meshing of realistic MRI RF coils with a high-resolution human model and electrodes results in a significant increase in computation time.
Different subgridding approaches are used to overcome this limitation of time-domain solvers. However, these are not very effective for MRI-related simulations due to the geometrical complexity of RF coils for MRI and different types of human tissue, the bent shape of electrodes, and the bent trajectories of the wires.
The aforementioned simulation drawbacks of voxel-based human models can be avoided with anatomically correct surface-based models and solvers based on unstructured meshes. A flexible discretization of human structures can be achieved with tetrahedra, pyramids, and extruded triangles (prisms) as mesh elements. In this way, the complex shape of curved human tissue structures, electrodes, and wires can be maintained. Electrical contact within and between tissues, as well as between an electrode and human tissue, can also be preserved.
For solvers based on unstructured grids, it is beneficial to have only one boundary between adjacent structures. When these boundaries are triangulated, the resulting surfaces are free from intersections or intermediate gaps, and the number of triangles in the entire model is reduced.
Unfortunately, 3D EM frequency-domain solver development has advanced beyond geometry import, pre-processing, and mesh generation capabilities. For most up-to-date solvers based on unstructured meshes, a surface-based model must only include objects that are geometrically error free (no self-intersections, over-connections, etc.), and the number of faces in the model must be limited to approximately 500,000 to be meshed by commercially available packages in an acceptable time interval.
Although the surface-based Virtual Family v2.x models [18] were developed primarily for 3D EM simulations, these models and high-resolution voxel-based human models (less than ~2 mm voxel size) do not meet the aforementioned error-free geometry requirements. Thus, their use with most up-to-date solvers based on unstructured meshes is practically impossible.
Two workhorses for 3D EM investigations are the Virtual Family v1.x models [19] and the Virtual Population 3.0 models [18]. Developed as surface-based anatomical models, they are used in 3D EM simulations as discretized voxel-based geometries. The Virtual Population 3.0 models are integrated within the multiphysics simulation platform Sim4Life, which includes only a time-domain EM solver, or the SEMCAD time-domain EM solver. The Virtual Population 3.0 models cannot be exported to any third-party software. The Virtual Family v1.x models are also compatible with Sim4Life or SEMCAD time-domain EM solvers or can be exported to other solvers only in voxel format, which is not suitable for import into solvers based on unstructured meshes.
Some of the surface-based human body models presented in 3D EM simulation reports, for example, the Chinese reference man [20], have been used only with FDTD solvers. The reasons for this are unknown.
Recent literature includes reports of the development of surface-based models for a variety of applications, for example, emission imaging (namely the 4-D XCAT Phantoms) [21], biomechanics, and injury biomechanics [22]. These application-specific models require an efficient conversion into a format that is compatible with a geometric modeling kernel of a 3D EM solver based on unstructured meshes or its geometrical pre-processor, as well as handling the geometrical complexity of the models at the appropriate level if a geometrical pre-processor cannot be used. The complexity of the direct conversion of an application-specific surface-based geometry to 3D EM suitable surface-based geometry could be a reason why a model should be voxelized as the first step, and new surface meshes should be generated as the second step, as was the case for 4-D XCAT Phantoms [23].
Converting voxel-based data to high-quality surface-based objects and correctly matching contact regions presents a significant challenge. It is even more difficult to meet all the requirements for importing a human model composed of numerous tissue structures into an EM solver in the form of surface-based geometries.
Only a few surface-based full-body human models, for example the NEVA Electromagnetics (Yarmouth Port, Cape Cod, MA, USA) female VHP model [24], developed based on the Visual Human Project® data set [25], and the Aarkid (East Lothian, Scotland) male model [26], have been used successfully with 3D EM solvers based on unstructured meshes. Available models provide different levels of detail of different human tissue types. For example, CSF is rarely included, and there are sometimes multiple levels of model fidelity.
The electrical properties of some types of human tissue are quite similar. Thus, a human model that only includes a subset of human tissue could be sufficient for application-specific MRI EM simulations. For MRI birdcage coil simulations, fat, muscle, bone, and air spaces are especially important to consider [26]. For high-field MRI head coil simulations, a human model should additionally include CSF, white matter (WM), and grey matter (GM) [27].
Generating a correct full-body surface-based model requires great effort throughout each stage. This is why head and torso models such as that developed by the team from NeuroSpin-CEA [28] have become effective solutions for investigating head RF exposure.
We previously introduced a semi-automatic processing pipeline to generate individualized surface-based models of the human head and upper torso from the MR images of individual subjects [29]. A key feature of this workflow is that the resulting models have a single surface between adjacent structures. The comprehensive workflow covers image acquisition, atlas-based segmentation of relevant structures, generation of segmentation masks, and surface mesh generation of the single, external boundary of each structure of interest. Two head and torso models were generated and used for 3D EM simulations using this pipeline [30].
The voxel models derived from the Visual Human Project Visible Man and Visible Woman data sets have formed the basis for a large number of MRI RF safety assessments [27]. See [31] for an example of the HUGO anatomical model. For interlab studies in general, it is beneficial to use voxel- and surface-based models derived from the same dataset. Therefore, we have selected the Visible Man data set as source data for this investigation.
In our case, the generated human models were intended for simulations of head coils in high-field MRI and 3T MRI whole body coils with patients at the head landmark position. For these purposes, a human model can be truncated at the torso without introducing substantial uncertainty.
In this investigation, the pre-segmented AustinMan dataset [32] was used to facilitate fast generation of the surface-based head and torso model of the Visual Human Project, Visible Man.
MRI coil development and the MRI RF safety assessment of a given RF coil require multi-port simulations, and results for only a single frequency in which the MRI scanner is running. The latter eliminates one of the major drawbacks of most frequency-domain solvers—the requirement to simulate a set of frequencies over the bands of interest. The size of the smallest mesh elements do not substantially influence the simulation time of most frequency-domain solvers. Thus, a frequency-domain solver is a good candidate for reliable RF safety assessment in MRI.
ANSYS HFSS (ANSYS, Inc., Canonsburg, PA, USA) was chosen for our 3D EM simulations because of its robustness in handling complex MRI coil geometries and fast multi-port simulations. Therefore, a substantial part of our work was to investigate optimization approaches that ensure successful 3D EM simulations when using surface-based geometry. It is important to note that the geometry kernel and associated functionality vary from solver to solver. Thus, some additional geometrical pre-processing may be required if our head and torso models are used with other 3D EM solvers.
The ANSYS Non-Linear Thermal (NLT) platform will be used for our future investigations into temperature rise for multimodal setups. Thus, the requirements of the ANSYS NLT platform were taken into account during development of the 3D EM model.
2 Methods
2.1 Surface Mesh Generation
Here, we present a dedicated subset of our previously established workflow, namely the post-processing of segmentation images to so-called segmentation masks followed by surface mesh generation. We applied this sub-part of the pipeline to the segmented AustinMan dataset [30].
We utilized the Medical Image Processing, Analysis and Visualization (MIPAV) toolset [33] (v.7.3) in conjunction with the Java Image Science Toolkit (JIST) [34] (v.2.0-2013) to automate the segmentation mask generation of the subsequently described procedures. The final surface meshing was realized in ParaView (v.5.0.1, Kitware Inc., New York, USA).
The AustinMan dataset was provided as a set of individual, segmented slices in the MATLAB MAT-file format . We did not select all available slices for further processing. Slices below the bottom of the lungs were discarded to generate a model of the head and upper torso. The in-slice resolution was three times higher than the resolution between slices, yielding a voxel size of 0.33 × 0.33 × 1.0 mm3. Using MATLAB, we converted the slices to a single volume image in the NIfTI file format, which can be imported into JIST. The structures as represented in the segmented image are unsuitable for surface meshing for several reasons. First, the anisotropic voxel size of the volume leads to an unbalanced level of detail in the three spatial directions. For this reason, the volume must be resampled to an isotropic voxel size. Second, due to their nested arrangement, most structures of the human body exhibit an outer and an inner boundary. However, the inner boundary may resemble the shape of the outer boundary of an adjacent internal structure. If triangulated, these adjacent boundaries are prone to mutual intersections and small gaps, which must be avoided. Third, the number and type of segmented structures exceed the typical level of detail required for EM simulations and can therefore be reduced. The segmented image is post-processed to segmentation masks to account for these requirements.
Using MIPAV, we resampled the image to an isotropic voxel size of 1 mm by reducing the in-slice resolution. We then split the image at a slice located at the chin of the enclosing exterior structure of the head to account for different requirements concerning the type of represented structures and different topological constraints of nested structures in the head and torso. The labels of these two images were integrated into a reduced set of labels comprising only the structures we aimed to consider for our EM simulations, namely enclosing exterior structure, bone, cerebrospinal fluid, the ventricles, cerebral GM and WM, the eyes, fat tissue only of the torso, muscle, and air. With the exception of the vascular system, all remaining structures, for example, the intestines, that could not be clearly assigned to one of these target structures were combined with the class of the muscle. The voxels representing the blood vessels had to be handled differently since the vascular system runs through many structures of the body. Assigning them to a class of musculature would introduce considerable and unreasonable segmentation errors, for example, muscle tissue inside the skull, the bones, or cerebrospinal fluid. Therefore, we cleared the labels of voxels representing the vascular system, that is, we assigned them the background value of 0. This procedure created holes in several structures at locations which were formerly attributed to the large draining veins of the brain. These holes were subsequently closed while the segmentation masks were being created. Finally, each structure was transferred to a separate image file and binarized.
Following these preparations, the segmentation masks for the head and body structures were generated separately. The images comprising the binarized, segmented structures were processed sequentially in a fixed order distinct for the body and the head. Separating the workflow for the head and torso was necessary since different topological constraints apply in each region. For example, air is entirely surrounded by bone in the sinuses of the skull, whereas the air in the lungs is outside any boney structure in the body.
The procedure started with the image representing the innermost structure, for example, the ventricles in the head, to the image representing the outermost structure, namely the enclosing exterior structure. Each image was processed identically: A morphological closing operation followed by a filling operation ensured a continuous outer boundary and eliminated the inner boundary as well as the holes created by the removal of the blood vessels. As a consequence, voxels that had originally been identified as blood vessels now represented the structure these vessels perfused. Small groups of detached voxels (namely less than 100 connected voxels) were identified as connected components and removed to obtain one large object per structure. A morphologically dilated version of the adjacent internal structure was added to the current structure. This way, we ensured a minimum thickness of two voxels for each structure surrounding another structure and avoided intersecting structures. An adapted approach was necessary in two cases: 1) Certain adjacent structures which were not nested still shared a common boundary (e.g., the thorax and the air in the lungs). In these cases the dilated mask image of one structure was subtracted from the mask image of the other, which created a spacing of at least two voxels between both structures. 2) The GM segmentation of AustinMan features very narrow sulci, down to the size of only one voxel, creating small detached islands of sulcal CSF in the GM. To avoid creating a discontinuous surface representation of the CSF structure, we applied a 2D filling operation to the GM mask for every slice independently, thereby eliminating such narrow sulci. The entire process resulted in individual segmentation masks for each tissue class of the head and torso separately, each with only a single external boundary.
The segmentation masks were then imported into ParaView. ParaView provides the so-called Contour Filter to compute a triangulated, polygonal representation of isosurfaces (namely surfaces of identical values in a 3D volume). The Contour Filter implements the “synchronized templates” algorithm [35], an improved version of the Marching Cubes algorithm [36]. As a result, we obtained high-resolution surfaces with a high number of triangles, which closely resembled the outer boundary of the segmentation masks, and the typical voxel grid-like structure of naїve surface reconstructions of structures from voxel-based 3D images was mitigated. These surfaces were then exported to individual files using the stereolithography (STL) format for subsequent processing.
The triangle size of the surface meshes had a side length of approximately 1 mm. This was defined by the resolution of the segmentation masks that ensured: (i) correct geometrical representation of inter-cranial tissues, and (ii) generated surface meshes to be geometrically error-free (no self-intersections, over-connection, etc.). The total number of faces was approximately 10 million, thus the human model that was generated with ParaView was unsuitable for simulation with ANSYS HFSS.
Depending on the resolution and the number of structures that are to be included in the model, the automated processing of the workflow took about 45 minutes to 1 hour. An additional 30 minutes of manual preparation was required by a trained and experienced person, for example, to combine some classes of tissue and for splitting the segmentation at the head.
2.2 Head and Torso Model Mesh Optimization for 3D EM Simulation in ANSYS HFSS
A human model in ANSYS HFSS must be represented as a set of solid bodies. ANSYS SpaceClaim (ANSYS, Inc., Canonsburg, PA, USA) was used as the geometry preprocessor for: (a) importing STL files, (b) verifying (and, if necessary, correction) that all surface meshes were error free (watertight, no disconnected regions, no self-intersections, no over-connections, etc.), (c) combining the head and torso sections of the exterior structure, (d) optimizing the number of mesh elements for each individual object, (e) converting surface meshes into solid bodies, and (f) exporting each object of the model in ACIS binary format, which is the native file format of the ANSYS HFSS geometric modeling kernel.
Steps “a” to “c” were implemented using the built-in functionality of the ANSYS SpaceClaim Faceted Data Toolkit. The Faceted Data Toolkit’s mesh repair functionality was sufficient for correcting a small number of mesh errors, but it was not suitable for handling the large number of mesh errors that appeared for most objects of our model if surface meshes contained triangles with a side length of more than 1 mm in ParaView. The main reasons for this were: (i) some compartments of the human model object were too thin (less than 3 mm) and it reduced the degree of freedom for correct mesh modification because of the low number of triangles and (ii) ParaView mesh errors were cascaded (disconnected regions, self-intersections, over-connections, etc.) in these areas.
Two approaches to reduce the number of mesh elements were applied in SpaceClaim: (i) All neighboring triangular faces located on the same geometrical plane were combined into a single face, and (ii) the number of faces were reduced by generating a triangular faceted wrapper around each model object.
An approach based on generating nonuniform rational B-spline (NURBS) surfaces was not implemented because the ANSYS HFSS geometry kernel operates internally with geometric primitives, for example, different types of facets and tetrahedra. Our tests based on ANSYS HFSS provided strong evidence that the meshing time for surface-based models based on NURBS is significantly longer than the time required for models generated using the previously mentioned approaches.
We also did not apply the ANSYS SpaceClaim Reduce tool to reduce the number of facets in a faceted body, because (i) for objects with a small number of facets, it provided a small reduction of the facet if spatial deviation was set to zero, (ii) mesh errors often resulted from complex geometric objects with a large number of facets.
2.2.1 Combining All Neighboring Triangular Faces
The “synchronized templates” algorithm generated high-resolution surface meshes that preserved all details of the underlying segmentation masks. Equally sized triangles were used for surface triangulations in the algorithm. This resulted in a redundant number of triangles, especially for large flat areas, because the 3D ACIS geometric modeling kernel (Dassault Systèmes, Vélizy-Villacoublay, France) does not require explicit triangulation to represent a flat area.
ANSYS SpaceClaim can combine all neighboring triangular faces located in the same geometrical plane into one face. The face reduction factor of this approach depends on the size of a given planar surface. For large planar surfaces, for example, those that are repeatedly represented on the enclosing-exterior-structure object, the reduction factor was very high (more than 100) (Fig. 13.1). For relatively small areas commonly observed in bent objects, for example, WM (Fig. 13.2), it was small (on the order of 10). This approach resulted in zero deviation of derived surface meshes from the original geometry.
The enclosing exterior structure object of AustinMan (a) after mesh generation with ParaView and (b) after combining faces. Head section of the skin object of AustinMan (c) after mesh generation and (d) after combining faces. Close-up view of the enclosing exterior structure object of AustinMan (e) after mesh generation and (f) after combining faces
Although the same 3D ACIS geometry modeling kernel is employed in both ANSYS SpaceClaim and ANSYS HFSS, different behavior was observed for the geometry validation check in ANSYS HFSS and ANSYS SpaceClaim if the face-combining approach was applied. The ANSYS HFSS geometry check reported errors for the objects that were error-free surface meshes in ANSYS SpaceClaim. One reason for this HFSS error is that the ANSYS SpaceClaim face combining procedure produces coincident edges that do not mark the boundaries of new faces. All ends that defined the combined faces are coincident edges. Therefore, the number of coincident edges is quite large, more than 10,000, for most model objects.
Our comprehensive ANSYS SpaceClaim tests of different human model geometries provided strong evidence that the ANSYS SpaceClaim Split Edges tool can detect and successfully merge coincident edges only if the number of coincident edges is relatively small, that is, less than approximately 1000.
Exporting human model objects prepared using ANSYS SpaceClaim to ACIS binary files was fast and problem free.
2.2.2 Faceted Wrapper
Using a second approach, generating a faceted wrapper around each model object decreased the number of faces. The reduction factors that specified the ratio of number of faces in the original object to the faceted wrapper object varied from approximately 5 to 40, depending on tissue importance for 3D EM simulations and geometrical complexity (Table 13.1).
The largest ratio was applied for the enclosing exterior structure (Fig. 13.3). This resulted in a deviation of up to 3 mm in the ear area. The smallest ratio was applied for WM, which resulted in a deviation of less than 0.2 mm between the original mesh and the wrapper (Fig. 13.4).
Generating a faceted wrapper around each model object did not result in geometry errors when the model was imported into ANSYS HFSS. Thus, the ANSYS HFSS healing procedure was not required for these model objects.
2.2.3 Comparison of Different Tissues
The shapes of the CSF, skull, and ribcage after geometric preprocessing for both approaches are shown in Figs. 13.5, 13.6, and 13.7.
Similar human model mesh optimization for 3D EM simulation in ANSYS HFSS was applied to prepare human Models 1 and 2 (Fig. 13.8) from surface meshes developed in our previous study [30]. One problem with most MRI scanners is that the maximum field of view is only 50 cm wide, and the patient on the patient table can only be moved in the axial direction, thus the subjects’ arms and shoulders can be truncated in the image data if a default imaging protocol is applied. This problem is most noticeable in Model 2.
2.3 A Test of Entire Body Mesh Optimization for 3D EM Simulation in ANSYS HFSS
To investigate the performance and limitations of our workflow for entire body model generation, the enclosing exterior structure object of the AustinMan model was generated using a segmentation mask resolution of 2 mm (Fig. 13.9a), while the extents and resolution of all other structures remained the same as in the head and torso model. The enclosing exterior structure object of the entire body resulted in 1,530,456 facets. The number of faces was reduced to 618,737 facets using a face-combining operation (Fig. 13.9b). The reduction ratio for a surface mesh where the side of a triangle was 2 mm was substantially smaller than for a surface mesh in which the side of a triangle was 1 mm. Use of a faceted wrapper for the enclosing exterior structure object of the entire AustinMan model with the same settings as the faceted wrapper for the enclosing exterior structure object of the AustinMan model’s head and torso resulted in significant spatial modification of areas between the model’s body and arms (Fig. 13.9c).
2.4 Finalizing the HFSS Model
Importing ACIS binary files exported from ANSYS SpaceClaim was fast and problem free in ANSYS HFSS. After import, each object underwent the ANSYS HFSS geometry validation check. A healing procedure was automatically applied if the ANSYS HFSS geometry check reported errors. This was a time-consuming process and took up to 10 hours to eliminate geometric errors per model object for complicated geometries if the face-combining approach was applied. ANSYS HFSS was not able to generate an error-free object after several days of healing the enclosing exterior structure of AustinMan.
An imported and healed (if necessary) model in ANSYS HFSS consisted of (i) the enclosing exterior structure, (ii) objects located above the chin slice created in the surface mesh generation step, primary head objects, and (iii) objects located below the chin slice, as well as primary torso objects.
The faceted wrapper slightly modified the geometries of model objects. However, an ANSYS HFSS simulation of a human model can consist of objects with both geometrical preprocessing approaches, because (i) deviation of any faceted wrapper from the original geometry is less than a quarter of the thickness of the given object, (ii) only one boundary between adjacent structures exists in areas outside the chin slice, and (iii) the intersection of adjacent structures in the chin slice area can be eliminated according to requirements.
To prevent intersections of objects in the area of the chin slice, all objects except the enclosing exterior structure underwent a boolean “split” operation in ANSYS HFSS. If the split operation resulted in two objects, only the primary object located above (for head objects) or below (for torso objects) the chin slice split plane was kept in the numerical domain.
2.5 Human Model Electrical Properties
Electrical properties of tissues were adopted from the IT’IS database [37]. Electrical property maps for electrical conductivity and relative electrical constant at 297.2 MHz provide a reasonable representation of human structures (Figs. 13.10, 13.11, and 13.12).
2.6 7T MRI Application-Specific Case Study
We performed 3D EM simulations of dual-row 7T head transmit array coil loaded with either the AustinMan model or Model 1 in ANSYS HFSS to evaluate the impact of human models on the spatially averaged 10-gram specific absorption rate (SAR10g), which is used as the RF power deposition safety limit in 7T head MRI transmission and safety efficiencies. The coil consisted of 16 identical rectangular loops (100 × 102.25 mm2) arranged in two rows of eight elements each (Fig. 13.13) [38]. A gap of 10 mm was applied between elements that were in the same row as well as between the two rows. The lower row elements were rotated by 22.5° with respect to the upper row. All adjacent elements were inductively decoupled.
The 3D EM model of the array included: (a) all array construction details for the resonant elements, (b) the load, namely, the surface-based human model, (c) the array environment, including the MRI scanner’s gradient shield and magnet bore, all simulated with precise dimensions and material electrical properties, and (d) inductive decoupling of all adjacent elements. However, neither RF cable traps nor coax cable interconnection wiring were included in the model.
Twelve distributed capacitors were inserted in each radiative element to provide feed, tune, shunt, and distributed capacitor functionality. One PIN diode with a resistance of 0.18 Ω was placed in series with one of the distributed capacitors. This diode was used for decoupling transmit-only radiative elements during MRI signal reception.
The decoupling networks were defined by inductors, with inductance Lind and coupling factor Kind, placed in series with the distributed capacitors. The Q factor of all capacitors was set to 324, and the Q factor of all inductors was set equal to 400.
The coil was tuned, matched, and decoupled for the single tissue phantom with an external shape like a human model [38]. The optimization of the transmitter coil was based on the minimization of an error function (EF), which was a measure of the difference between the actual and desired coil conditions. Commonly used criteria for multi-channel RF transmitters, at the desired frequency, are: (a) the element reflection coefficient Sxx must be set and equal to a required value (i.e., Sxx_t) for each coil element, and (b) the element coupling between adjacent elements Sxy must be equal to a required value (i.e., Sxy_t) for each decoupled element pair. Hence
where Elem is the number of loops of the coil (namely 16) and all_dec is the number of decoupled element pairs (namely 32).
Both rows were excited in circular polarization (CP) mode with phase difference φrow of 22.5° between rows. RF circuit and 3D EM co-simulation as detailed in [39] was used for calculations.
SAR10g was calculated using an in-house procedure, which is consistent with the IEEE/IEC 62704-1 standard and validated by means of an IEEE TC 34 interlab comparison study [40].
2.7 3T MRI Application-Specific Case Study
The 3D EM model of the whole-body coil utilized a 123.2 MHz 16-rung high-pass birdcage of an equivalent design to those widely used in clinical 3T scanners (inner diameter 615 mm, total length 480 mm). The model head was positioned at the isocenter of the coil (Fig. 13.14a). The coil was shielded by a metal enclosure that mimicked a 1220 mm-long scanner bore. To mimic the scanner room, the coil was centered in an air box with the dimensions of 3 × 2.25 × 5 m3, surrounded by perfectly matched layer boundaries on all sides. The coil was tuned, matched, and decoupled using an elliptical phantom (length 700 mm, major radius 175 mm, minor radius 95 mm) positioned in the isocenter of the coil. The phantom material properties were: electrical conductivity σ = 0.52 S/m and relative permittivity εr = 53.4 (Fig. 13.14b). The optimization procedure, RF circuit and 3D EM co-simulation for the 3T birdcage coil were similar to the 7T transmit coil simulations described in the previous section.
The amplitude of the two RF sources used to excite the coil was the same for both feeds, with a 90° phase shift between the feeds as in quadrature excitation. All results were calculated for a transmission power of 2 W.
The NEVA Electromagnetics VHP high-resolution whole human model [24] was used to check our assumption that the head and torso model is sufficient for 3T investigations of the head landmark position (Fig. 13.14c). The electrical properties of different types of human tissue were adopted from the IT’IS database [37].
2.8 RF Safety of Transcranial Direct Current Stimulation Equipment During MRI Case Studies
The impedance of electrical contact between an electrode and the skin should be low, e.g. during tDCS or EEG procedures. A conductive gel is used to minimize this impedance, which must be included in the numerical domain because it modifies the RF field in the proximity of the electrode. Placement of a gel patch between the electrode and the skin surface (Fig. 13.15a) resulted in small faces on the edges of the patch (Fig. 13.15b). Such small faces could complicate the generation of high quality numerical meshes, for example, in the ANSYS Non-Linear Thermal (NLT) platform.
Therefore, the triangular faces of the skin object in areas around the electrode were merged into a single NURBS face (Fig. 13.16a). Designed using the native geometrical capability of the ACIS kernel in ANSYS HFSS, the electrode and gel patch were located at the required positions in close proximity to the skin (Fig. 13.16b). After boolean subtraction of the gel object by the skin object, a correct single face contact between the gel object and skin object was obtained (Fig. 13.16c).
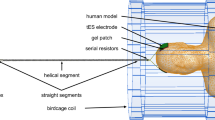
The tDCS setup consisted of two electrodes, two leads, and a metal connection box located 410 mm away from the coil enclosure. A composite-material quadratic tDCS electrode was simulated as a conductive medium with εr = 3 and σ = 4 S/m. The serial resistors integrated in the leads were located 100 mm away from the electrodes. Three resistor values were simulated: 1 mΩ, 5 kΩ, and 1 GΩ to simulate conditions of a short (potential manufacturing fault), normal operation, and an open connection (resistor failure after long-term operation), respectively. The tDCS lead included several straight segments and one helical segment. The lead copper wire was 1.2 mm in diameter with an insulation of 2.2 mm diameter. The helix pitch was 12.5 mm. Two tDCS lead trajectories were simulated: first on the axis of the scanner bore (recommended in the device manual) and then shifted towards the edge of the patient table.
3 Numerical Simulation Results
3.1 7T MRI Coil Simulation Results
The coil appeared to be correctly tuned for a given load with Sxx (the element reflection coefficient) values of less than −30 dB and Sxy (the element coupling between adjacent elements) values of less than −16 dB (Fig. 13.17).
CSF acted as a weak RF screen (Figs. 13.18 and 13.19), resulting in: (i) a decrease of B1+ at the skull/CSF boundary at the top part of the scalp, (ii) a substantial drop of the magnetic transmit field, B1+, in GM and WM, and (iii) a significant redistribution of volume loss density. Concomitantly, power deposition increased in the CSF space (Fig. 13.20). Visible variation of B1+ and volume loss density was observed for the investigated models.
Changing the human model resulted in some variation of B1+ and SAR10g profiles (Figs. 13.21 and 13.22). Additionally, the transmission efficiency and the safety excitation efficiency were higher for Model 1.
3.2 3T MRI Coil Simulation Results
Circuit-level optimization resulted in an appropriately tuned birdcage coil with an elliptical phantom present in the bore (Fig. 13.23a). Unsurprisingly, the S parameters were visibly affected when the coil was loaded with human models at the head landmark position, which resulted in asymmetrical coil loading (Fig. 13.23b). No substantial difference in S parameters were observed for the head and torso of the AustinMan model or the NEVA Electromagnetics VHP entire human model.
For both human models, 3D EM results were consistent with common observations in the literature, for example [41]: B1+ was rather homogeneously distributed across the head, and the maximum deposition of power occurred in the neck region (Figs. 13.24 and 13.25). As for the 7T coil simulation, if CSF was represented in the numerical domain as a non-separated, continuous segment within a space, it acted as a weak RF screen resulting in: (i) a decrease of B1+ at the skull/CSF boundary at the top part of the scalp and (ii) a significant volume loss density in CSF.
Truncation of the AustinMan model at the torso did not significantly affect the birdcage coil circuit level results or field distributions. Only a very weak scattered field was observed in the area located in close proximity to the torso cut plane.
3.3 Transcranial Direct-Current Stimulation Results
After adding the tDCS setup with the lead directed along the magnet axis, substantial power deposition was observed in close proximity to the tDCS electrode edges for all values of the serial resistor. Unsurprisingly, the B1+ disturbance in close proximity to the electrode location was highest for R = 1 mΩ. For a normal tDCS setup operation with R = 5 kΩ, shifting the tDCS lead from the scanner axis toward the edge of the patient table resulted in a small variation of power deposition in the proximity of the electrode edges. Assuming a (pulsed) peak value of 30 kW of the total transmission power (which can be generated by the scanner’s power amplifier) yielded voltages across the serial resistor up to 850 V for normal tDCS operation (R = 5kΩ) and up to 1.4 kV for an open connection (R = 1 GΩ). For a whole-body SAR level of 4 W/kg, average voltages across the serial resistor were 130 V and 225 V for R = 5 kΩ and R = 1 GΩ, respectively. The obtained range of voltages underscores how sufficient electrical strength (e.g., order of 1 kV) is required for the tDCS serial resistor. Due to the similarity of the power deposition in the proximity of the electrode edges for all investigated conditions, we conclude that the tDCS electrodes and the straight segments of the leads between them and the serial resistor predominantly determine the power deposition in human subjects.
4 Discussion
Our investigation explored the impact of patient-specific human models on MRI safety assessment from different perspectives. Future work should address how many different human models, head positions, and non-ideal tuning conditions need to be investigated and how many different excitation conditions need to be validated in order to demonstrate MRI RF transmit coil robustness, as well as MRI multimodal setup and implant RF safety.
Our mesh optimization procedure for the 3D EM simulation workflow is specifically tailored toward performing simulations with ANSYS HFSS and ANSYS NLT. Use of other simulation tools could require some modification of geometry preparation steps, for example, the generation of NURBS surfaces instead of faceted objects.
In our previous work, we have introduced a semi-automated processing pipeline to generate individualized surface-based models from MRI data of individual subjects. While this pipeline offers a high level of automation, especially concerning the segmentation of the MRI data and segmentation mask generation, so far it is limited to model a few relevant structures (i.e., the enclosing exterior structure, bone, air, GM, WM, and CSF). Limitations mainly arise from difficulties in segmenting certain inter-subject variable tissue types in MR images.
MRI data provides good contrast for different types of soft tissue, but additional effort is required to segment skin and bone, especially when this should be achieved in an algorithm-driven manner, without supervision of an expert. Fully unsupervised segmentation of highly variable structures, for example, muscle and fat tissue, from MRI data across subjects is challenging to achieve using our atlas-based approaches and is therefore still subject to further research. However, if corresponding segmentation images were available, our segmentation mask generation workflow could be extended to include these additional structures, as detailed in this work.
To prevent geometrical model errors in most simulation tools and to accelerate geometrical export and preprocessing, our segmentation mask generation process enforces the topological constraint that adjacent structures should not share a common boundary. The segmented structures were modified according to topological constraints for the human anatomy: (i) to being either strictly nested or (ii) not in contact with boundaries of neighboring structures. As a result, for example, the ventricles of the brain are entirely surrounded by WM, which again is fully surrounded by GM even at the brain stem, and there is a space between the rib cage and the lung object.
The more structures that are represented in the model, the more difficult it becomes to maintain this topological constraint. For example, the vascular system runs through a major subset of all the other structures, which made it impossible to fully nest it inside another single structure. Furthermore, introducing a space between its boundary and the boundaries of all the other structures would create holes in those structures.
Additionally, some tissue segments were too small to be represented, which, for example, was the case for the pieces of CSF in some narrow sulci in the brain. We therefore opted for an approach that eliminates the CSF in these sulci to ensure a continuous boundary for the subarachnoid CSF, resulting in trade-off of a less accurate representation of sulcal CSF.
As a consequence of both aforementioned problems, we did not include fat tissue in the head region. More specifically, fat tissue in the head is present in several types of tissue, for example, skin and muscle tissue. As a result, the fat exhibits common boundaries with several other structures, such as the skull, cartilage tissue, tendon tissue, and the eyes, which made it impossible to entirely nest it inside one structure. Additionally, the fat tissue was not segmented in a continuous way, larger gaps existed that could not be closed with morphological closing operations and some segments of fat tissue were as thin as only one voxel. A possible solution to address these obstacles might be to divide the class of fat tissue into subclasses for which compliance to the topological constraints can be achieved more easily. We are working on defining a set of rules on how to reasonably combine the mentioned classes of tissue in an informed anatomical way, and how to handle the discontinuous fat tissue and thereby ensure compliance with the necessary topological constraints.
In this work, we have elaborated on the necessary workflow using the AustinMan model. However, we expect our workflow to also work for other segmented data sets, such as NAOMI [42] and NORMAN [43] and voxel model databases of the average Japanese male and female [44]. If segmentation images are already available for a person from a previous investigation, our segmentation mask generation workflow can be applied to generate a surface-based head and torso model for this individual. Depending on the quality and continuity of the segmented structures, adaptations will only be necessary with regard to the integration of the available tissue classes into the desired set of structures, the order of structures for which the segmentation mask generation will be executed, from the innermost to the outermost structure, and the position of where to split the head section of the segmentation image from the torso section. These adjustments can be achieved in a time frame of approximately 1 day.
In addition, for these new models it is important to investigate whether certain structures need dedicated treatment, for example, as was observed in the narrow sulci of the GM, the vascular system, or the fat tissue in the head of the AustinMan model. Resolving these special cases may require adaptations as simple as adjusting the parameters for morphological operations (i.e., closing or filling), which was the case for the narrow sulci in the GM. Alternatively, they may require a dedicated sub-workflow to be developed, which was the case for the vascular system, and which would be the case for handling fat tissue in the head of the AustinMan. In the latter case, the necessary time frame of adapting the proposed workflow may easily increase to several days.
An extension of the presented workflow to create whole-body models will be the next step. We expect similar difficulties with body fat, as we discovered for the head and limbs, especially in the abdominal region where the intestines are located.
The time required for geometry modification, import, preprocessing, and mesh generation was ten times longer than the solver time of approximately 2 hours on an up-to-date Dell workstation. This is not compatible with real-time patient-specific safety assessment. However, it is reasonable for investigating more realistic distributions of human body shapes and sizes to explore the variation of SAR values between subjects, as well as SAR dependences on intracranial geometric variation (e.g., variation of CSF spaces with age).
Further development of ANSYS SpaceClaim and ANSYS HFSS capabilities: (i) to reduce the amount of facets in surface meshes without creating geometrical problems in ANSYS HFSS, and (ii) fast geometry import, preprocessing, and mesh generation for geometries with a large number of facets in ANSYS HFSS, could substantially decrease the time needed for 3D EM simulation of high-resolution human models.
References
International Electrotechnical Commission (IEC). (2010). Medical electrical equipment-part 2–33: Particular requirements for the basic safety and essential performance of magnetic resonance equipment for medical diagnosis. Geneva, Switzerland: International Electrotechnical Commission, 60601-2-33 Ed. 3.
Shajan, G., Kozlov, M., Hoffmann, J., Turner, R., Scheffler, K., & Pohmann, R. (2014). A 16-channel dual-row transmit array in combination with a 31-element receive array for human brain imaging at 9.4 T. Magnetic Resonance in Medicine, 71(2), 870–879.
Oh, S., Webb, A. G., Neuberger, T., Park, B., & Collins, C. M. (2010). Experimental and numerical assessment of MRI-induced temperature change and SAR distributions in phantoms and in vivo. Magnetic Resonance in Medicine, 63, 218–223.
Murbach, M., Neufeld, E., Cabot, E., Zastrow, E., Córcoles, J., Kainz, W., & Kuster, N. (2016). Virtual population-based assessment of the impact of 3 Tesla radiofrequency shimming and thermoregulation on safety and B1+ uniformity. Magnetic Resonance in Medicine, 76(3), 986–997.
Murbach, M., Neufeld, E., Kainz, W., Pruessmann, K. P., & Kuster, N. (2014). Wholebody and local RF absorption in human models as a function of anatomy and position within 1.5T MR body coil. Magnetic Resonance in Medicine, 71, 839–845.
Voigt, T., Homann, H., Katscher, U., & Doessel, O. (2012). Patient-individual local SAR determination: In vivo measurements and numerical validation. Magnetic Resonance in Medicine, 68, 1117–1126.
Wu, X., Tian, J., Schmitter, S., Vaughan, J. T., Uǧurbil, K., & Van De Moortele, P.-F. (2016). Distributing coil elements in three dimensions enhances parallel transmission multiband RF performance: A simulation study in the human brain at 7 Tesla. Magnetic Resonance in Medicine, 75(6), 2464–2472.
Ryan, K., Wawrzyn, K., Gati, J. S., Chronik, B. A., Wong, D., Duggal, N., & Bartha, R. (2018). 1H MR spectroscopy of the motor cortex immediately following transcranial direct current stimulation at 7 Tesla. PLoS One, 13(8). Article number e0198053.
Lee, M. B., Kim, H. J., Woo, E. J., & Kwon, O. I. (2018). Anisotropic conductivity tensor imaging for transcranial direct current stimulation (tDCS) using magnetic resonance diffusion tensor imaging (MR-DTI). PLoS One, 13(5). Article number e0197063.
Keinänen, T., Rytky, S., Korhonen, V., Huotari, N., Nikkinen, J., Tervonen, O., Palva, J. M., & Kiviniemi, V. (2018). Fluctuations of the EEG-fMRI correlation reflect intrinsic strength of functional connectivity in default mode network. Journal of Neuroscience Research, 96(10), 1689–1698.
Bailey, W., Mazur, A., McCotter, C., Woodard, P.K., Rosenthal, L., Johnson, W., & Mela, T. (2016). Clinical safety of the ProMRI pacemaker system in patients subjected to thoracic spine and cardiac 1.5-T magnetic resonance imaging scanning conditions. Heart Rhythm, 13(2), 464–471.
Bhusal, B., Bhattacharyya, P., Baig, T., Jones, S., & Martens, M. (2018). Measurements and simulation of RF heating of implanted stereo-electroencephalography electrodes during MR scans. Magnetic Resonance in Medicine, 80(4), 1676–1685.
Guerin, B., Serano, P., Iacono, M.I., Herrington, T.M., Widge, A.S., Dougherty, D.D., Bonmassar, G., Angelone, L.M., & Wald, L.L. (2018). Realistic modeling of deep brain stimulation implants for electromagnetic MRI safety studies”, Physics in Medicine and Biology, 63(9), Article number 095015.
Atefi, S. R., Serano, P., Poulsen, C., Angelone, L. M., & Bonmassar, G. (2018). Numerical and experimental analysis of radiofrequency-induced heating versus lead conductivity during EEG-MRI at 3 T. IEEE Transactions on Electromagnetic Compatibility, (99). https://doi.org/10.1109/TEMC.2018.2840050.
Kozlov, M., & Kainz, W. (2018). Lead electromagnetic model to evaluate RF-induced heating of a coax lead: A numerical case study at 128 MHz. IEEE Journal of Electromagnetics, RF and Microwaves in Medicine and Biology. https://doi.org/10.1109/JERM.2018.2865459.
ASTM F2182-11a. (2011). Standard test method for measurement of radio frequency induced heating on or near passive implants during magnetic resonance imaging. West Conshohocken, PA: ASTM International, www.astm.org.
Xu, X. G. (2014). An exponential growth of computational phantom research in radiation protection, imaging, and radiotherapy: A review of the fifty-year history. Physics in Medicine and Biology, 59, R233–R302.
Gosselin, M.-C., Neufeld, E., Moser, H., Huber, E., Farcito, S., Gerber, L., Jedensjo, M., Hilber, I., Gennaro, F.D., Lloyd, B., Cherubini, E., Szczerba, D., Kainz, W., & Kuster, N. (2014). Development of a new generation of high-resolution anatomical models for medical device evaluation: The virtual population 3.0. Phys. Med. Biol., 59(18), 5287–5303.
Christ, A., Kainz, W., Hahn, E.G., Honegger, K., Zefferer, M., Neufeld, E., Rascher, W., Janka, R., Bautz, W., Chen, J., Kiefer, B., Schmitt, P., Hollenbach, H.-P., Shen, J., Oberle, M., Szczerba, D., Kam, A., Guag, J.W., & Kuster, N. (2010). The virtual family—Development of surface-based anatomical models of two adults and two children for dosimetric simulations. Physics in Medicine and Biology, 55(2), 23–38.
Yu, D., Wang, M., & Liu, Q. (2015). Development of Chinese reference man deformable surface phantom and its application to the influence of physique on electromagnetic dosimetry. Physics in Medicine and Biology, 60(17), 6833–6846.
Segars, W.P., Tsui, B.M.W., Cai, J., Yin, F.-F., Fung, G.S.K., & Samei, E. (2018). Application of the 4-D XCAT phantoms in biomedical imaging and beyond. IEEE Transactions on Medical Imaging, 37(3), 680–692.
Elemance: The sole distributor of the global human body models consortium family of virtual models of the human body, [online] Available: http://www.elemance.com/
Genc, K.O., Segars, P., Cockram, S., Thompson, D., Horner, M., Cotton, R., & Young, P. (2013). Workflow for creating a simulation ready virtual population for finite element modeling. Journal of Medical Devices, 7(4), 1–2. https://doi.org/10.1115/1.4025847.
Makarov, S. N., Noetscher, G. M., Yanamadala, J., Piazza, M. W., Louie, S., Prokop, A., Nazarian, A., & Nummenmaa, A. (2017). Virtual human models for electromagnetic studies and their applications. IEEE Reviews in Biomedical Engineering, 10, 95–121. http://ieeexplore.ieee.org/document/7964701/.
Spitzer, V., Ackerman, M. J., Scherzinger, A. L., & Whitlock, D. (1996). The visible human male: A technical report. Journal of the American Medical Informatics Association: JAMIA, 3, 118–130.
Homann, H., Börnert, P., Eggers, H., Nehrke, K., Dössel, O., & Graesslin, I. (2011). Toward individualized SAR models and in vivo validation. Magnetic Resonance in Medicine, 66, 1767–1776.
Kozlov, M., Bazin, P.-L., Möller, H. E., & Weiskopf, N. (2016). Influence of cerebrospinal fluid on specific absorption rate generated by 300 MHz MRI transmit array. Proceedings of 10th European Conference on Antennas and Propagation (EuCAP). https://doi.org/10.1109/EuCAP.2016.7481666.
Massire, A., Cloos, M.A., Luong, M., Amadon, A., Vignaud, A., Wiggins, C.J., & Boulant, N. (2012). Thermal simulations in the human head for high field MRI using parallel transmission. J. Magn. Reson. Imag., 35(6), 1312–1321.
Kalloch, B., Bode, J., Kozlov, M., Pampel, A., Hlawitschka, M., Sehm, B., Villringer, A., Möller, H. E., & Bazin, P.‐L. (2019). Semi‐automated generation of individual computational models of the human head and torso from MR images. Magnetic Resonance in Medicine, 81(3), 2090–2105.
Kozlov M., Bode J., Bazin P.-L., Kalloch B., Weiskopf N., Moeller H.E. (2017). Building a high resolution surface-based human head and torso model for evaluation of specific absorption rates in MRI (pp. 1–6), Proceedings of COMCAS 2017. Tel-Aviv, Isreal.
Gjonaj, E., Bartsch, M., Clemens, M., Schupp, S., & Weiland, T. (2002). High-resolution human anatomy models for advanced electromagnetic field computations. IEEE Transactions on Magnetics, 38(2), 357–360.
Massey, J. W., & Yilmaz A. E. (2016). AustinMan and AustinWoman: high-fidelity, anatomical voxel models developed from the VHP color images. In Proc. 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (IEEE EMBC). Orlando, FL.
Mcauliffe, M., Lalonde, F., McGarry, D. P., Gandler, W., Csaky, K., Trus, B. (2001). Medical image processing, analysis & visualization in clinical research. In Proceedings of the 14th IEEE Symposium on Computer-Based Medical Systems (pp. 381–386).
Lucas, B. C., Bogovic, J. A., Carass, A., Bazin, P. L., Prince, J. L., Pham, D. L., & Landman, B. A. (2010). The Java Image Science Toolkit (JIST) for rapid prototyping and publishing of neuroimaging software. Neuroinformatics, 8, 5–17.
Moreland, K., & Geveci, B. (2014). A pervasive parallel processing framework for data visualization and analysis at extreme scale. Clifton Park, NY/Albuquerque, NM: Tech. rep. Kitware, Inc./Sandia National Lab.(SNL-NM).
Lorensen, W. E., Cline, H. E. (1987). Marching cubes: a high resolution 3D surface construction algorithm. In Proceedings of the 14th annual conference on computer graphics and interactive techniques (pp. 163–169).
Hasgall, P. A., Di Gennaro, F., Baumgartner, C., Neufeld, E., Gosselin, M. C., Payne, D., Klingenböck, A., Kuster, N. (2015). IT’IS database for thermal and electromagnetic parameters of biological tissues, Version 3.0, September 1. www.itis.ethz.ch/database
Kozlov, M., Turner, R., Weiskopf, N., Möller, H. E., Shajan, G. (2017). Investigation of 7T 16-channel dual-row transmit array coils: a case study of static RF shimming. In Proceedings of COMCAS 2017 (pp. 1–5). Tel Aviv.
Kozlov, M., & Turner, R. (2009). Fast MRI coil analysis based on 3-D electromagnetic and RF circuit co-simulation. Journal of Magnetic Resonance, 200, 147–152.
Monebhurrun, V., Braux, Y., Devos, H., Kozlov, M., Simon, W., & Wittig, T. (2014). A benchmark CAD mobile phone model for specific absorption rate calculations. IEEE Transactions on Magnetics, 50(2). https://doi.org/10.1109/TMAG.2013.2282470.
Yeo, D., Wang, Z., Loew, W., Vogel, M., & Hancu, I. (2011). Local SAR in high pass birdcage and TEM body coils for multiple human body models in clinical landmark positions at 3T. Journal of Magnetic Resonance Imaging, 33(5), 1209–1217. https://doi.org/10.1002/jmri.22544.
Dimbylow, P. (2005). Development of the female voxel phantom, NAOMI, and its application to calculations of induced current densities and electric fields from applied low frequency magnetic and electric fields. Physics in Medicine and Biology, 50(6), 1047–1070. https://doi.org/10.1088/0031-9155/50/6/002.
Findlay, R. P., & Dimbylow, P. J. (2009). Spatial averaging of fields from half-wave dipole antennas and corresponding SAR calculations in the NORMAN human voxel model between 65 MHz and 2 GHz. Physics in Medicine and Biology, 54(8), 2437–2447. https://doi.org/10.1088/0031-9155/54/8/012.
Nagaoka, T., Watanabe, S., Sakurai, K., Kunieda, E., Watanabe, S., Taki, M., & Yamanaka, Y. (2004). Development of realistic high-resolution whole-body voxel models of Japanese adult male and female of average height and weight, and application of models to radio-frequency electromagnetic-field dosimetry. Physics in Medicine and Biology, 49, 1–15.
Acknowledgments
The authors acknowledge the Computational Electromagnetics Group at the University of Texas at Austin for developing and making the AustinMan human body model available at http://sites.utexas.edu/austinmanaustinwomanmodels/. The authors also acknowledge the ANSYS Partnership Program for providing the numerical simulation software for these investigations.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2019 The Author(s)
About this chapter
Cite this chapter
Kozlov, M., Kalloch, B., Horner, M., Bazin, PL., Weiskopf, N., Möller, H.E. (2019). Patient-Specific RF Safety Assessment in MRI: Progress in Creating Surface-Based Human Head and Shoulder Models. In: Makarov, S., Horner, M., Noetscher, G. (eds) Brain and Human Body Modeling. Springer, Cham. https://doi.org/10.1007/978-3-030-21293-3_13
Download citation
DOI: https://doi.org/10.1007/978-3-030-21293-3_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-21292-6
Online ISBN: 978-3-030-21293-3
eBook Packages: EngineeringEngineering (R0)