Published online Dec 10, 2015. doi: 10.5306/wjco.v6.i6.252
Peer-review started: May 9, 2014
First decision: July 11, 2014
Revised: September 3, 2015
Accepted: October 1, 2015
Article in press: October 8, 2015
Published online: December 10, 2015
Breast cancer is an intrinsically heterogeneous disease. In the world about 1 million cases of breast cancer are diagnosed annually and more than 170000 are triple-negative. Characteristic feature of triple negative breast cancer (TNBC) is that it lacks expression of oestrogen, progesterone and human epidermal growth factor receptor-2/neu receptors. They comprise 15%-20% of all breast cancers. We did a systematic review of PubMed and conference databases to identify studies published on biomarkers in TNBC. We included studies with biomarkers including: Epidermal growth factor receptor, vascular endothelial growth factor, c-Myc, C-kit and basal cytokeratins, Poly(ADP-ribose) polymerase-1, p53, tyrosinase kinases, m-TOR, heat and shock proteins and TOP-2A in TNBC. We also looked for studies published on synthetic lethality and inhibition of angiogenesis, growth, and survival pathways. TNBC is a complex disease subtype with many subclasses. Majority TNBC have a basal-like molecular phenotype by gene expression profiling. Their clinical and pathologic features overlap with hereditary BRCA1 related breast cancers. Management of these tumours is a challenge to the clinician because of its aggressive behaviour, poor outcome, and absence of targeted therapies. As the complexity of this disease is being simplified over time new targets are also being discovered for the treatment of this disease. There are many biomarkers in TNBC being used in clinical practice. Biomarkers may be useful as prognostic or predictive indicators as well as suggest possible targets for novel therapies. Many targeted agents are being studied for treatment of TNBC.
Core tip: Triple negative breast cancer (TNBC) are type of breast cancer which lack of estrogen receptors, progesterone receptors and human epidermal growth factor receptor. It is a complex disease subtype with many subclasses. There are many biomarkers in TNBC used for its sub-classification. Clinically-practical assay/biomarkers that can reliably identify TNBC are necessary. Biomarkers may be useful as prognostic or predictive indicators as well as suggest possible targets for novel therapies.
- Citation: Yadav BS, Chanana P, Jhamb S. Biomarkers in triple negative breast cancer: A review. World J Clin Oncol 2015; 6(6): 252-263
- URL: https://www.wjgnet.com/2218-4333/full/v6/i6/252.htm
- DOI: https://dx.doi.org/10.5306/wjco.v6.i6.252
Breast cancer is a complex disease entity with different biological characteristics and clinical behaviour. Many clinical and pathological features have been defined to predict outcome and treatment response in breast cancer. These features include: Patient age, tumour stage, axillary lymphnode involvement, lymphovascular invasion, histologic grade, hormonal and human epidermal growth factor receptor (HER-2/neu receptor) status. In the past chemotherapy was the only systemic therapy for triple negative breast cancer (TNBC) patients. Currently lot of research is going on to further characterise TNBC with different molecular markers and find targets for therapy in order to improve its outcome. Sørlie et al[1] has diversified five subgroups of breast cancer by gene expression profiling (GEP) using DNA microarrays. These are luminal A, luminal B, HER-2/neu over expressing, basal like (BL) and normal like breast cancer. BL breast cancer lacks estrogen receptors (ER), progesterone receptors (PR) and HER-2/neu receptors, thus contribute to 80% of TNBC[1,2] The present review provides an insight into the different biomarkers in TNBC and its sub classification based upon the marker profile to understand molecular targets in each subtype.
TNBC[3] are type of breast cancer which lack ER, PR and HER-2/neu receptors. It has different and poor clinical and pathological features as compared to other subtypes of breast cancer. It is usually seen in young age, advanced stage at presentation, unfavourable histopathology, grade III, higher proliferative index, lack of tubule formation and higher rate of metastases[4-9]. It is associated with higher rate of local recurrence during 3 year after treatment and a high 5 year death rate[10]. Survival is poor after distant metastasis[11,12]. TNBC frequently affects younger patients (< 50 years) and has higher prevalence in the African-American women[13]. Patients with TNBC has inferior disease free survival (DFS) and overall survival (OS) as compared to age and grade matched controls of non-TNBC patients[11]. In TNBC metastatic rate is high to visceral organs[14,15] and lung and cerebral metastasis is more common[16-19]. Cytotoxic chemotherapy is the only treatment option[20-22].
TNBC is a distinct breast cancer. It is classified into six groups based upon the GEP and DNA microarray. This sub-classification is not only useful in understanding the disease better but also to find molecular targets for its treatment[23].
BL-1 and BL-2: The BL-1 subtype was found to be composed rapidly dividing cells associated with increased proliferation and cell cycle checkpoint loss consistent with the increased expression of DNA damage response genes. Due to its high proliferation rate it has increased Ki67 mRNA expression and it is more responsiveness to antimitotic agents targeting cell cycle. The BL-2 subtype on the other hand displayed unique gene ontologies involving epidermal growth factor signalling as well as glycolysis and gluconeogenesis pathway. On microarray it showed a higher expression of epidermal growth factor receptor (EGFR), TP63, MET, etc.
Immunomodulatory subtype: Immunomodulatory (IM) is composed of immune cell responses such as immune cell and cytokine signalling, antigen presentation and processing and signalling of immune transduction pathways. Its GEP substantially overlaps with the medullary breast cancer, histologically a rare distinct form of TNBC which carry favourable prognosis despite its high grade.
Mesenchymal and mesenchymal stem like subtype: On GEP these subtypes consists of epithelial-mesenchymal (M) transition and growth factor pathways. The mesenchymal stem like subtype is also expressed by genes involved in angiogenesis including VEGFR2 and was found to be highly responsive to dasatinib [tyrosine kinase (TK) inhibitor], and mTOR inhibitors.
Luminal androgen receptor subtype: This subtype is characterised by androgen receptor (AR) signalling. It is ER negative but gene ontologies were heavily composed of hormonally regulated pathways such as steroid synthesis, porphyrin metabolism and androgen/estrogen metabolism. AR mRNA expression was nine times higher than other subtypes therefore, these lines were found to be highly sensitive to AR antagonists eg biclutamide. Patients with this subtype had decreased DFS and OS.
Among TNBCs 80%-90% falls into the category of BL molecular subtype when appropriately tested for IHC cancer biomarkers and GEP but these terms are nonsynonymous and are overlapping[10,24]. At present, there is no optimal IHC panel for identification of basal like breast cancer (BLBC). Therefore TNBC, despite having above limitations is considered as a BL cancer. In a study Thike et al[9] with a tri-panel of cytokeratin-14 (CK-14) , EGFR and 34βE12 in TNBC reported 84% to be BL tumors with a specificity and sensitivity of 100% and 78% respectively. In BLBC over expression of ID4 leads to the deregulation of BRCA1. BLBCs are also known to have either p53 over expression or mutations in the gene[24].
In array, BLBCs are characterised by low expression of ER and HER-2 related genes, so pathologically they are usually ER-negative, PR-negative and lack HER-2 over expression[8,9] or are < 1%; < 5%; 10%; 20% immunoreactive for the above receptors[24]. They stains positive for cytokeratins (CKs) 5/6 and 17, and over express EGFR (HER1). Furthermore they show a highly aggressive GEP with low Bcl-2 but high p53 and Ki67[25-29].
Genetic instability leads to cancer predisposition. Genetic mutations in the BRCA genes in patients predisposes them to develop many cancers such as breast, ovarian, pancreatic and prostate. BRCA 1 plays vital role in DNA repair by homologous recombination. Inactivation of this gene due to BRCA mutation should trigger cell cycle arrest but this too is inhibited by p53 mutations in TNBC[30]. Lack of a functional BRCA1/2 in cells lead to loss of repair of DNA double-strand breaks (DSB). This mechanism leads to increased risk of cancer in these patients. Histologically and transcriptionally, TNBC share similarities with BRCA1-linked breast cancers, which means that dysfunction of BRCA1 is seen in TNBCs[31,32]. TNBCs are heterogeneous with respect to GEP. TNBC is associated with cancers arising in BRCA1 mutation carrier in young women as compared to those in their late forties. Both sporadic BLBCs and BRCA1 associated breast cancers have evidence of genomic instability. More than 80% of breast cancers in women who carry germ-line BRCA1 mutations are TN and 10% TN breast tumors have BRCA1 mutation. The reasons for these associations are unclear but may ultimately provide avenues for prevention as well as targeted therapy with poly(ADP-ribose) polymerase (PARP) inhibitors and chemotherapy with DNA-damaging agents such as platinum compounds[33-35].
TNBC is characterised by the marked expression of certain biomarkers. The presence of these molecules though is not restricted to TNBC but somehow show increased prevalence in this subgroup. The following are the important biomarkers in TNBC.
EGFR: EGFR is one of the members of four closely related receptors each playing an important role in tumour cell survival. The four receptors being EGFR (or ErbB-1), HER-2/neu (ErbB-2), HER-3 (ErbB-3), and HER-4 (ErbB-4)[36,37]. The inactive monomer receptor dimerizes after ligand activation followed by TK, intracellular domain of the receptor is activated by autophosphorylation, leading to cascade of intracellular events. EGFR signal cascade is important for cell proliferation, angiogenesis, metastatic spread, and the inhibition of apoptosis[38]. Most of the TNBCs express EGFR, and poses a strong therapeutic challenge[39]. Studies with different methods of gene amplification have found variable expression EGFR in metaplastic breast carcinoma, a phenotypes of BLBCs[40-42]. However, Toyama et al[43] with real-time polymerase chain reaction have reported high EGFR gene copy number in TNBCs. EGFR expression is found in 40%-50% of patients with breast cancer and in 80% of TNBC; and is estimated to substitute major proliferation pathways of breast cancer induced by activation of HER-2, ER, PR proteins which are thereby absent in TNBC[25].
In a study the authors found that 60% of patients with grade III and > 3 lymph nodes showed EGFR expression, indicating that EGFR expression is related to aggressiveness of the disease. They also concluded that patients with EGFR expression had worse DFS, distant disease free survival (DDFS), OS and cause specific survival[44]. EGFR expression in TNBC is associated with poor response to chemotherapy[45]. Nogi et al[46] observed that EGFR was expressed in 24% of the TNBC patients and was related to less favourable response to chemotherapy and poorer survival and on the contrary the luminal groups where EGFR expression showed good response to chemotherapy and better survival. Recently EGFR has been defined with other markers to differentiate BL subtype from TNBC[47]. This aids in segregating TNBC into subtypes and thus defining the prognostic difference and molecular target specification between the two. Non-uniformity of expression profiles in studies shown in Table 1 is due to absence of subtype consideration or BL subtype non segregation from core TNBC. So EGFR is a biomarker in TNBC and a target for cetuximab, a TK inhibitor[48]. Many studies have evaluated its response in TNBC[48-51]. In a recent study, EGFR expression was shown as prognostic factor for DFS not only in univariate but also in multivariate analysis[52].
| Ref. | Total number | No. of TNBC subjects | EGFR expression1 |
| Thike et al[9], 2010 | 7048 | 767 | 30% |
| Patil et al[10], 2011 | 683 | 136 | 7.4% |
| Nielsen et al[24], 2004 | - | 21 basal like tumours | 57% |
| Rakha et al[45], 2007 | 1726 | 282 | 37% in TNBC vs 15% in non-TNBC |
| Mehdizadeh et al[47], 2012 | 1132 | 103 | 23.3% |
| Rydén et al[48], 2010 | 564 | 48 | 41% TNBC vs 11% non-TNBC |
Vascular endothelial growth factor: Angiogenesis is important for tumour growth and spread especially beyond a diameter of 2 mm as oxygen and nutrients cannot diffuse beyond this distance. Angiogenic signals are mediated by vascular endothelial growth factor (VEGF) to aid neovascularisation. VEGF A, B, C, D, E (viral factor) and placental growth factor is a family of six proteins. VEGF protein is found in 4 isoforms because of alternative splicing of its mRNA[53,54]. Among the different isoforms VEGF165, the 165-amino acid molecule is more common[55,56]. Its gene expression is controlled by many of stimuli such as hypoxia, nitric oxide, growth factors, oncogenes, tumour suppressor genes and HER-2[57].
It causes proliferation and maintains structural and functional integrity of cells of the endothelium. It also regulates vascular permeability and migration of endothelial stem cells from the bone marrow[58]. Neovascularisation in the tumour is also regulated by VEGF by increasing the expression of the anti-apoptotic proteins such as Bcl2, XIAP, and survivin. In its absence the endothelial cells undergo apoptosis and newly formed vessels disintegrate[59-61]. Thus neovascularisation is dependent on VEGF expression throughout tumour development. VEGF shows multiple interactions with receptor TKs, such as VEGFR-1, VEGFR-2, and VEGFR-3. The angiogenesis is initiated by VEGF binding to VEGFR-2 which triggers the specific activation of TKs followed by multiple signalling cascades resulting in the endothelial cells survival, proliferation, migration, adhesion, actin remodelling and vessels permeability[62].
VEGF expression is elevated in DCIS and invasive breast cancer. It has been also well utilised for prognosis in breast cancer[63,64]. Its quantification by IHC or immunoassay of tissue extracts has shown a significant co relation with micro vessels counts or density. High mean vascular density in breast cancer has been found to linked with more aggressive tumour behaviour and poor survival so intratumoral microvessels density is now considered as one of the important factors affecting survival[65]. According to recent studies[63,66] there was a direct co relation between serum and tissue levels of VEGF to grade III tumours, larger tumour size, positive lymph node and negative hormone status and poor survival along with a substantial decrease in levels with chemotherapy. In TNBC higher VEGF levels are associated with shorter DFS, OS, and DDFS. Also VEGF levels have been significantly related to size of the tumour, grade and metastatic sites. In patients with higher VEGF levels disease progressed despite of therapy and such patients were associated with significantly lower progression free survival as compared to patients with lower levels. In TNBC patients it was found that VEGF level elevated from baseline to middle of the therapy significantly but showed a non significant increase from middle of the therapy to its end when patients were administered FAC[65-67]. VEGF is a target for bevacizumab in TNBC patients. Table 2 shows VEGF expression reported in different studies.
C-kit and basal cytokeratins: C-kit is a cytokine receptor present on the surface of hematopoietic stem cells and also in other cells. C-kit binds to stem cell factor and is a growth factor receptor that stimulates major cellular functions such as cell survival, proliferation, differentiation, adhesion and chemotaxis. It induces apoptosis and also increases the invasiveness of the cancer cells[68]. CKs are keratin-containing proteins of intermediate filaments found in the intracytoplasmic cytoskeleton of epithelial tissue. Different epithelial tissues express different CKs at the time of its terminal differentiation and the stage of development. This different CK expression helps in the classification of all epithelia. Similarly different cancers express specific CKs of that epithelium. Therefore the CK expression profile tends to remain constant when an epithelium undergoes malignant transformation.
The study of the CK profile by IHC techniques is very important for tumor pathologic classification[69]. These CKs were earlier used to distinguish malignant breast lesions from benign ones[70], but later their prognostic value was ascertained and it was seen that expression of CK-5, CK-14 and CK-17 was related to poor prognosis, high grade tumours, ER negativity, short DFS and OS[71-73]. It is expressed in BLBCs. Since BLBC and TNBC show overlapping features therefore C-kit and basal CKs along with other markers and pathological features are used for the differentiating BLBCs from TNBC. Many studies have revealed that presence of CKs is higher in TNBC than non-TNBC and also among TNBC subgroup it is higher in the BL subclass (Table 3). BL subclass of TNBC was identified on the basis of CK and EGFR expression and when the clinicopathological features were compared between the basal and non-BL it was seen that BL subclass of TNBC were more aggressive[9,74-78].
p53: It is a tumour suppressor protein which is encoded by the TP53 gene (the tumour suppressor gene). It is also called the “guardian of genome” as it is important cell cycle regulator[79]. It regulates cell growth, multiplication, proliferation and apoptosis, and promotes chromosomal stability. Disruption of these functions by mutation in the gene producing p53 lead to carcinogenesis. p53 is activated in response to cellular stress by many pathways that are dependent on distinct upstream regulatory kinases. First, an ataxia-telangectasia mutated proteins released in response to the DSB, second, a pathway dependent on INK4 gene product, p14ARF activated by oncogenes, and finally, a pathway induced by chemotherapy drugs and ultraviolet light and is independent of the above two pathways[80,81].
p53 mutations are seen in 18%-25% of primary breast carcinomas (Table 4)[82]. p53 plays an important role in breast cancer prognosis. p53 over expression leads to poor response to chemotherapy[83,84]. Many studies have reported that its activation is associated with aggressive form of breast cancer and significantly decreases DFS and OS in TNBC patients[85-88]. Also co existence with HER-2 was significantly related to early relapse and death within shorter period after surgery[87]. Along with EGFR and cytokeratins it is used for segregation of a subclass, i.e., basal like from core TNBC[89].
| Ref. | Total number | No. of TNBC | p-53 expression |
| Patil et al[10], 2011 | 683 | 135/683 | 47.8% |
| Nielsen et al[24], 2004 | 11 | 11 | 82% |
| Rakha et al[45], 2007 | 1726 | 282/1726 | 56% in TNBC vs 22% in non-TNBC |
| Chae et al[90], 2008 | 135 | 32/135 | 40.6% in TNBC vs 42.7% in non- TNBC |
| Biganzoli et al[89], 2011 | - | (633 + 1026) from two separate sources | Divided TNBC into subclass BL which accounts for 89% of total TNBCs |
Tumours with p53 mutation are highly invasive, poorly differentiated and high grade tumours. In a study by Chae et al[90], p53 mutation was associated with poor response to the chemotherapy in TNBC patients. Other proteins of p53 family are p63/p73 proteins. Tumors expressing these proteins are reported to have many folds higher sensitivity to platinum based chemotherapy. p63/p73 expression is seen in one-third of patients with TNBC[91].
TOP-2A: This gene encodes topoisomerase II α and plays a crucial role in DNA transcription. This enzyme causes the temporary break of double strands of duplex DNA and rejoins them so that the strands cross through one another, therefore altering the topology of DNA. Mutation in cancer leads to depreviation of its functions and thus worsening of the situation. In TNBC or breast carcinoma the gene acts as a target for anthracycline therapy which is a topoisomerase II inhibitor[92]. So it is a marker for the evaluation of resistance to the anthracycline therapy. A study revealed a higher expression of TOP-2A in 2.7% to 8.8% of TNBC patients[93]. Its over expression in TNBC leads to the decreased sensitivity towards the anthracyclines and thus decreased response[94].
Ki67: Also known as MKI67, Ki67 is a cellular marker for proliferation. Ki67 antigen is present inside the cell nucleus during interphase and during mitosis it is relocated to the surface of the chromosomes. Since it is a marker of proliferation it is found in all cells when they are in dividing phases of the cell cycle (G1, S, G2, and mitosis) and it is absent from cells during their resting phase (G0). Its absence in resting cells and generalised presence in dividing cells had made it a marker of cell proliferation[95]. Proliferation is a salient feature for the spread of cancer and can be assessed by the IHC measurement of the nuclear antigen Ki67. It’s over expression also correlates with levels of bromodeoxyuridine uptake and S-phase fraction, other markers of proliferation.
Ki67 expression is less in normal breast tissue (< 3%). It has been reported in many studies that Ki67 antigen and steroid-receptor are expressed in different cells in normal human breast epithelium. Ki67 was over expressed particularly in ER-negative cells and its expression in carcinoma cells was much higher[96,97]. In breast cancer high Ki67 is associated with of poor outcome although these tumours show very good clinical response to combination chemotherapy. However, its independent significance is modest and does not merit measurements in routine clinical practice. With respect to treatment response in breast cancer, Ki67 expression was found to be independent predictor of pathologic complete response (pCR), clinical complete response, OS and DDFS and locoregional recurrence. It was also seen that patients without pCR still showed a decrease in Ki67 index post therapy[98-100]. In a recent meta-analysis by de Azambuja et al[101] who retrieved DFS data from 29 studies, they concluded that high Ki67 levels was associated with poor prognosis in irrespective of nodal status and whether patients undergo treatment or not at all.
In TNBC, it was found that Ki67 levels were significantly increased in ductal TNBC compared to other histologic types (80% in TNBC vs 10%-30% in other types). Its expression also represented a direct co relation with tumour size and grade in TNBC patients and higher levels (> 35% staining) were linked with an increased risk of death[102,103]. In TNBC patients Ki67 accumulation was associated with a higher pCR to chemotherapy but poor RFS and OS. Its expression was also used for subdivision of TNBC into two subtypes where only 26.7% of TNBC patients showed lower Ki67 expression[104].
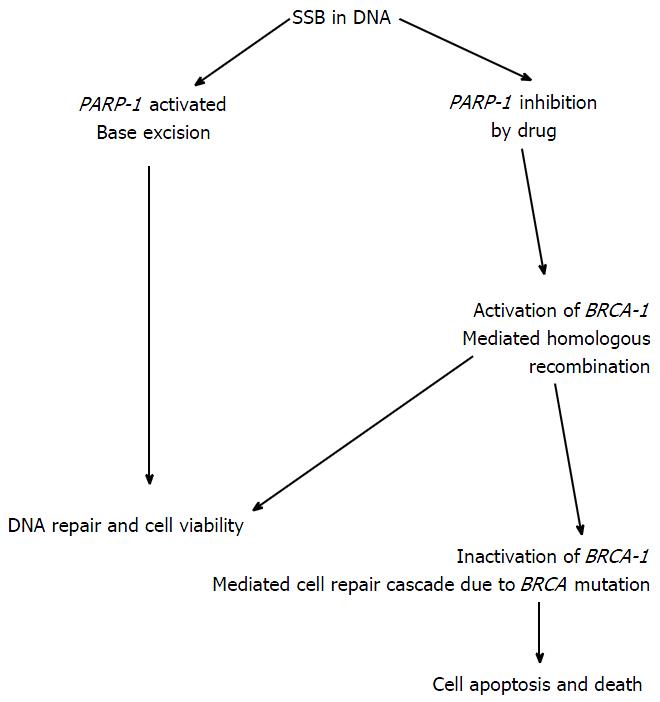
PARP: PARPs are a family of cell signalling enzymes present in eukaryotes, which catalyses the poly(ADP-ribosylation) of DNA binding proteins. Till now eighteen enzymes of PARPs has been detected, but PARP1 the most common isoform. PARP1 is responsible for majority of its functions. Main function of PARP1 is as DNA damage nick sensor. It forms polymers of ADP-ribose and nicotinamide with use of NAD+. Activation of PARP1 is important in tumours because of three interesting biological reasons: First, it plays a vital role in DNA repair through base excision repair pathway; second, it is capable of depleting cellular energetic pools, which results in cell dysfunction and necrosis; and third, its ability to promote the transcription of proinflammatory genes. PARP enzymes are involved in cellular response in inflammation, ischemia and oxidative stress. Carcinogenesis is a multistep process involving alterations in many cellular processes such as genomic stability, cell division, proliferation, growth, differentiation and cell death. PARP1 are involved in all these cellular processes, indicating possible link between PARP1 function and carcinognesis[105]. PARP1 repairs DNA single strand breaks (SSB) by binding to the exposed ends of the damaged DNA strand and bring in important enzymes required for repair in SSBs[106-110]. The base excision repair pathway fails when PARP1 is inhibited; this leads to accumulation of SSBs. In a dividing cell entering S-phase, cell division is arrested at SSBs, leading to a DSB (Figure 1). In BRCA1 deficient cells excision repair pathway is dependent on PARP1, inhibition of PARP1 leads to cell death through apoptosis[106,107]. BRCA2 operates through excision repair pathway like BRCA1, mutation of this gene make the cells suceptible to PARP inhibitors as well[109,110]. PARP also plays a vital role in DNA repair as BRCA. Unlike BRCA it recognises SSBs and repairs by base excision repair pathway[105]. PARP inhibitors are effective in TNBC because damage to one of the arms of the DNA could not be repaired by homologous recombination due to BRCA mutation and PARP inhibition in synergism will create a state of “synthetic lethality” - a process that occurs when inactivation of individual genes have no effect but mutations in both the genes lead to death of cancer cells[107]. So BRCA mutation is responsible for the action of many chemotherapeutic agents in TNBC. The inhibition of PARP1 is also known to potentiate the effect of ionizing radiation and many drugs such as DNA methylating agents, topoisomerase I inhibitors, and platinum compounds. Studies in mouse models have shown that the addition of PARP inhibitors with platinum compounds increases RFS and OS[35,105,107] while many of other studies on cell lines reveal that the activity of PARP inhibitors was increased in presence of BRCA mutations or dysfunction[105,108]. PARP1 has been targeted as therapeutic option in TNBC with drugs like iniparib, olaparib etc though not found to be independently helpful but their addition to cytotoxic agents have surely brought synergism to their activity and improvement in treatment response in TNBC patients.
Heat shock protein 90: It is a cellular chaperone (proteins that assist the assembly or disassembly of other macromolecular structures) protein that mediates the post-translational modification and stabilization of a number of conformationally labile proteins, steroid receptors, cyclin-dependent kinase 4, RAF-1, AKT and other proteins that are useful for sending proliferative signals[111]. Once function of heat shock protein (HSP) 90 is blocked, its dependent proteins are broken by proteosomes. Small HSP αB-crystalline is expressed in BLBCs and is associated with shorter survival. Its’ over expression is associated with neoplastic changes in mammary acini, increases cell migration and invasion in vitro. Geldanamicyn and tanespimycin both are antibiotics and inhibitors of HSP. These have shown clinical benefit in HER2-positive metastatic breast cancer[112]. The PU-H71 another HSP blocker has shown complete response in TNBC models[113].
Cox-2: Cox is a conversion enzyme of arachidonic acid and prostaglandin. It is a 74kDa protein located in the cell endothelium, reticulum and nuclear membrane. It is expressed by stimuli such as inflammatory response and tumor promoters. In a study by Liu et al[114] they observed that 85% of transgenic mice with over expression of Cox developed breast cancer, suggesting the involvement of this enzyme in breast carcinogenesis. Other studies have correlated its expression with invasiveness and metastatic stimuli in breast cancer[115,116]. Approximately 40% of patient with breast cancer over expresses Cox-2. Cox-2 can also be used as a biomarker to assess response to neoadjuvant chemotherapy in breast cancer.
Lymph node status is major of prognostic significance in breast cancer patients. Studies have shown that Cox-2 expression is associated with positive lymph node involvement. So Cox-2 may have some role in lymphangiogenesis. Cox-2 expression has been also correlated to hormone receptors in breast cancer, negative hormone receptors with Cox-2 expression indicate worse prognosis. Cox-2 is correlated to HER2 through Ras/MAPK pathway and it is associated with HER2 over expression[117]. Cox-2 expression is also related to MDR-1, a multidrug resistance gene. Patients with expression of both these are least responsive to chemotherapy. So Cox-2 can be a good biomarker in breast cancer patients with its correlation with size of the tumour, number of nodes involved, hormone receptors and HER2 status[118].
TK: TKs are regulatory proteins that help in the cell growth and differentiation. These proto-oncogenes play an important role in progression and metastasis of cancer cells. They also increase sensitivity of cancer cells once the tumour has been exposed to radiation and chemotherapy through apoptosis[36]. Hence, TKs are of major interest and are subject of many active studies to look targets for therapeutic intervention in many solid tumours. HER2/neu and EGFR are also TKs receptors as discussed above. HER2/neu over-expression is seen in 20%-25% of invasive breast cancers and it is considered a poor prognostic factor. Other TKs over-expressed in carcinoma of the breast are BRK, c-Src, and EGFR[119]. Lack of expression of some of TKs such as Syk and C-kit are also linked to carcinogenesis of breast cancer. TK over-expression in women with breast cancer is have high risk of metastasis. There are many agents that target the phosphorylation of the receptor by acting at TK[120]. TK inhibitors such as imatinib, erlotinib, gefitinib and lapatinib are used for treatment of many solid tumours. Dasatinib and lapatinib are used in treatment of women with HER2/neu positive breast cancer.
Mammalian target of rapamycin: One of the pathway is commonly dysregulated in breast cancer is phosphatidylinositol 3-kinase/mammalian target of rapamycin (PI3K/mTOR). Over expression of the PI3K/mTOR is associated with poor response to treatment with hormones and trastuzumab[121]. To overcome endocrine resistance agents such as rapalogs, that efficiently block mTOR-raptor complex 1, can be used along with hormones. However, it has demonstrated variable results in hormone receptor positive metastatic breast cancer[122].
Many targets such as αVβ6, cyclin E, C-kit, E-cadherin, O6MGMT, FOXp3, β-blockers, insulin like growth factors, glycoprotein NMB and mitogen-activated protein kinase pathway needs further exploration to dissect TNBC and may possibly identify new biomarkers and targets for therapy.
TNBC is the most poorly understood and is refractory to current targeted therapies. It is a cause of significant breast cancer mortality because of very few treatment options. Biomarker may be useful as prognostic or predictive indicators as well as suggest possible targets for novel therapies. Targeted therapy directed against many biomarkers has not shown significant improvement in outcome in TNBC, therefore it is challenging for the clinicians to deal with this distinct disease. The emphasis should be put on research for effective drugs and targets for the treatment TNBC. So, to translate the present knowledge about TNBC into oncological practice, biomarkers/molecules/GEP assays that can truly classify TNBC and can be easily translated to the clinics are necessary.
P- Reviewer: Camacho J, Langdon S
S- Editor: Ji FF L- Editor: A E- Editor: Li D
| 1. | Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869-10874. [PubMed] [Cited in This Article: ] |
| 2. | Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA. Molecular portraits of human breast tumours. Nature. 2000;406:747-752. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10665] [Cited by in F6Publishing: 10440] [Article Influence: 435.0] [Reference Citation Analysis (0)] |
| 3. | Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429-4434. [PubMed] [Cited in This Article: ] |
| 4. | Cleator S, Heller W, Coombes RC. Triple-negative breast cancer: therapeutic options. Lancet Oncol. 2007;8:235-244. [PubMed] [Cited in This Article: ] |
| 5. | Irvin WJ, Carey LA. What is triple-negative breast cancer? Eur J Cancer. 2008;44:2799-2805. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 207] [Cited by in F6Publishing: 223] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 6. | Stockmans G, Deraedt K, Wildiers H, Moerman P, Paridaens R. Triple-negative breast cancer. Curr Opin Oncol. 2008;20:614-620. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 51] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 7. | Reis-Filho JS, Tutt AN. Triple negative tumours: a critical review. Histopathology. 2008;52:108-118. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 594] [Cited by in F6Publishing: 614] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 8. | Diaz LK, Cryns VL, Symmans WF, Sneige N. Triple negative breast carcinoma and the basal phenotype: from expression profiling to clinical practice. Adv Anat Pathol. 2007;14:419-430. [PubMed] [Cited in This Article: ] |
| 9. | Thike AA, Cheok PY, Jara-Lazaro AR, Tan B, Tan P, Tan PH. Triple-negative breast cancer: clinicopathological characteristics and relationship with basal-like breast cancer. Mod Pathol. 2010;23:123-133. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 162] [Cited by in F6Publishing: 171] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 10. | Patil VW, Singhai R, Patil AV, Gurav PD. Triple-negative (ER, PgR, HER-2/neu) breast cancer in Indian women. Breast Cancer (Dove Med Press). 2011;3:9-19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 11. | Mersin H, Yildirim E, Berberoglu U, Gülben K. The prognostic importance of triple negative breast carcinoma. Breast. 2008;17:341-346. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 83] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 12. | Rodríguez-Pinilla SM, Sarrió D, Honrado E, Hardisson D, Calero F, Benitez J, Palacios J. Prognostic significance of basal-like phenotype and fascin expression in node-negative invasive breast carcinomas. Clin Cancer Res. 2006;12:1533-1539. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 262] [Cited by in F6Publishing: 257] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 13. | Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109:1721-1728. [PubMed] [Cited in This Article: ] |
| 14. | Smid M, Wang Y, Zhang Y, Sieuwerts AM, Yu J, Klijn JG, Foekens JA, Martens JW. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 2008;68:3108-3114. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 655] [Reference Citation Analysis (0)] |
| 15. | Lin NU, Claus E, Sohl J, Razzak AR, Arnaout A, Winer EP. Sites of distant recurrence and clinical outcomes in patients with metastatic triple-negative breast cancer: high incidence of central nervous system metastases. Cancer. 2008;113:2638-2645. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 505] [Cited by in F6Publishing: 490] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 16. | Heitz F, Harter P, Lueck HJ, Fissler-Eckhoff A, Lorenz-Salehi F, Scheil-Bertram S, Traut A, du Bois A. Triple-negative and HER2-overexpressing breast cancers exhibit an elevated risk and an earlier occurrence of cerebral metastases. Eur J Cancer. 2009;45:2792-2798. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 148] [Cited by in F6Publishing: 158] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 17. | Kaplan HG, Malmgren JA, Atwood M. T1N0 triple negative breast cancer: risk of recurrence and adjuvant chemotherapy. Breast J. 2009;15:454-460. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 76] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 18. | Haffty BG, Yang Q, Reiss M, Kearney T, Higgins SA, Weidhaas J, Harris L, Hait W, Toppmeyer D. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J Clin Oncol. 2006;24:5652-5657. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 767] [Cited by in F6Publishing: 803] [Article Influence: 44.6] [Reference Citation Analysis (0)] |
| 19. | Tsuda H, Takarabe T, Hasegawa F, Fukutomi T, Hirohashi S. Large, central acellular zones indicating myoepithelial tumor differentiation in high-grade invasive ductal carcinomas as markers of predisposition to lung and brain metastases. Am J Surg Pathol. 2000;24:197-202. [PubMed] [Cited in This Article: ] |
| 20. | Liedtke C, Mazouni C, Hess KR, André F, Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B, Green M. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275-1281. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1808] [Cited by in F6Publishing: 1999] [Article Influence: 124.9] [Reference Citation Analysis (0)] |
| 21. | Kassam F, Enright K, Dent R, Dranitsaris G, Myers J, Flynn C, Fralick M, Kumar R, Clemons M. Survival outcomes for patients with metastatic triple-negative breast cancer: implications for clinical practice and trial design. Clin Breast Cancer. 2009;9:29-33. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 282] [Cited by in F6Publishing: 308] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 22. | Rouzier R, Perou CM, Symmans WF, Ibrahim N, Cristofanilli M, Anderson K, Hess KR, Stec J, Ayers M, Wagner P. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11:5678-5685. [PubMed] [Cited in This Article: ] |
| 23. | Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750-2767. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3591] [Cited by in F6Publishing: 3617] [Article Influence: 278.2] [Reference Citation Analysis (0)] |
| 24. | Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367-5374. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1840] [Cited by in F6Publishing: 1849] [Article Influence: 97.3] [Reference Citation Analysis (0)] |
| 25. | Bidard FC, Conforti R, Boulet T, Michiels S, Delaloge S, André F. Does triple-negative phenotype accurately identify basal-like tumour? An immunohistochemical analysis based on 143 ‘triple-negative’ breast cancers. Ann Oncol. 2007;18:1285-1286. [PubMed] [Cited in This Article: ] |
| 26. | Livasy CA, Karaca G, Nanda R, Tretiakova MS, Olopade OI, Moore DT, Perou CM. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19:264-271. [PubMed] [Cited in This Article: ] |
| 27. | Banerjee S, Reis-Filho JS, Ashley S, Steele D, Ashworth A, Lakhani SR, Smith IE. Basal-like breast carcinomas: clinical outcome and response to chemotherapy. J Clin Pathol. 2006;59:729-735. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 215] [Cited by in F6Publishing: 207] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 28. | Osbourne CR, Kannan L, Ashfaq R, Ariyibi J, Frawley WH, Tripathy D. Clinical and pathological characterization of basal-like breast cancer. Breast Cancer Res Treat. 2005;118. [Cited in This Article: ] |
| 29. | Tischkowitz M, Brunet JS, Bégin LR, Huntsman DG, Cheang MC, Akslen LA, Nielsen TO, Foulkes WD. Use of immunohistochemical markers can refine prognosis in triple negative breast cancer. BMC Cancer. 2007;7:134. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 264] [Cited by in F6Publishing: 284] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 30. | Foulkes WD, Stefansson IM, Chappuis PO, Bégin LR, Goffin JR, Wong N, Trudel M, Akslen LA. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst. 2003;95:1482-1485. [PubMed] [Cited in This Article: ] |
| 31. | Lakhani SR, Reis-Filho JS, Fulford L, Penault-Llorca F, van der Vijver M, Parry S, Bishop T, Benitez J, Rivas C, Bignon YJ. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res. 2005;11:5175-5180. [PubMed] [Cited in This Article: ] |
| 32. | Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer. 2004;4:814-819. [PubMed] [Cited in This Article: ] |
| 33. | Quinn JE, Kennedy RD, Mullan PB, Gilmore PM, Carty M, Johnston PG, Harkin DP. BRCA1 functions as a differential modulator of chemotherapy-induced apoptosis. Cancer Res. 2003;63:6221-6228. [PubMed] [Cited in This Article: ] |
| 34. | Tassone P, Tagliaferri P, Perricelli A, Blotta S, Quaresima B, Martelli ML, Goel A, Barbieri V, Costanzo F, Boland CR. BRCA1 expression modulates chemosensitivity of BRCA1-defective HCC1937 human breast cancer cells. Br J Cancer. 2003;88:1285-1291. [PubMed] [Cited in This Article: ] |
| 35. | Rottenberg S, Jaspers JE, Kersbergen A, van der Burg E, Nygren AO, Zander SA, Derksen PW, de Bruin M, Zevenhoven J, Lau A. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci USA. 2008;105:17079-17084. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 661] [Cited by in F6Publishing: 723] [Article Influence: 45.2] [Reference Citation Analysis (0)] |
| 36. | Wells A. EGF receptor. Int J Biochem Cell Biol. 1999;31:637-643. [PubMed] [Cited in This Article: ] |
| 37. | Noonberg SB, Benz CC. Tyrosine kinase inhibitors targeted to the epidermal growth factor receptor subfamily: role as anticancer agents. Drugs. 2000;59:753-767. [PubMed] [Cited in This Article: ] |
| 38. | Siziopikou KP, Cobleigh M. The basal subtype of breast carcinomas may represent the group of breast tumors that could benefit from EGFR-targeted therapies. Breast. 2007;16:104-107. [PubMed] [Cited in This Article: ] |
| 39. | Bhargava R, Gerald WL, Li AR, Pan Q, Lal P, Ladanyi M, Chen B. EGFR gene amplification in breast cancer: correlation with epidermal growth factor receptor mRNA and protein expression and HER-2 status and absence of EGFR-activating mutations. Mod Pathol. 2005;18:1027-1033. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 222] [Cited by in F6Publishing: 235] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 40. | Reis-Filho JS, Pinheiro C, Lambros MB, Milanezi F, Carvalho S, Savage K, Simpson PT, Jones C, Swift S, Mackay A. EGFR amplification and lack of activating mutations in metaplastic breast carcinomas. J Pathol. 2006;209:445-453. [PubMed] [Cited in This Article: ] |
| 41. | Gilbert JA, Goetz MP, Reynolds CA, Ingle JN, Giordano KF, Suman VJ, Blair HE, Jenkins RB, Lingle WL, Reinholz MM. Molecular analysis of metaplastic breast carcinoma: high EGFR copy number via aneusomy. Mol Cancer Ther. 2008;7:944-951. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 56] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 42. | Gwin K, Lezon-Geyda K, Harris L, Tavassoli FA. Chromosome 7 aneusomy in metaplastic breast carcinomas with chondroid, squamous, and spindle-cell differentiation. Int J Surg Pathol. 2011;19:20-25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 43. | Toyama T, Yamashita H, Kondo N, Okuda K, Takahashi S, Sasaki H, Sugiura H, Iwase H, Fujii Y. Frequently increased epidermal growth factor receptor (EGFR) copy numbers and decreased BRCA1 mRNA expression in Japanese triple-negative breast cancers. BMC Cancer. 2008;8:309. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 58] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 44. | Viale G, Rotmensz N, Maisonneuve P, Bottiglieri L, Montagna E, Luini A, Veronesi P, Intra M, Torrisi R, Cardillo A. Invasive ductal carcinoma of the breast with the “triple-negative” phenotype: prognostic implications of EGFR immunoreactivity. Breast Cancer Res Treat. 2009;116:317-328. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 144] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 45. | Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO. Prognostic markers in triple-negative breast cancer. Cancer. 2007;109:25-32. [PubMed] [Cited in This Article: ] |
| 46. | Nogi H, Kobayashi T, Suzuki M, Tabei I, Kawase K, Toriumi Y, Fukushima H, Uchida K. EGFR as paradoxical predictor of chemosensitivity and outcome among triple-negative breast cancer. Oncol Rep. 2009;21:413-417. [PubMed] [Cited in This Article: ] |
| 47. | Mehdizadeh R, Nazafi S, Jahanjad I. Evaluation of EGFR, VEGFR2, IGF-1R, and HIF-1a expression and their prognostic value in Iranian triple negative breast cancer patients. EJC. 2012;48:S145. [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 48. | Rydén L, Jirström K, Haglund M, Stål O, Fernö M. Epidermal growth factor receptor and vascular endothelial growth factor receptor 2 are specific biomarkers in triple-negative breast cancer. Results from a controlled randomized trial with long-term follow-up. Breast Cancer Res Treat. 2010;120:491-498. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 49. | O’Shaughnessy J, Weckstein DJ, Vukelja SJ. Preliminary results of a randomized phase II study of weekly irinotecan/carboplatin with or without cetuximab in patients with metastatic breast cancer. Breast Cancer Res Treat. 2007;106:S32 Abstract 308. [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 57] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 50. | Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492-2502. [PubMed] [Cited in This Article: ] |
| 51. | Khambata-Ford S, O’Shaughnessy J, Brickman D, Jensen JD, Asmar L, Horak CE, Baylor NJ. Candidate predictive biomarkers of cetuximab benefit in triple negative breast cancer. J Clin Oncol. 2010;28:15s (suppl; abstr 1056). [Cited in This Article: ] |
| 52. | Liu D, He J, Yuan Z, Wang S, Peng R, Shi Y, Teng X, Qin T. EGFR expression correlates with decreased disease-free survival in triple-negative breast cancer: a retrospective analysis based on a tissue microarray. Med Oncol. 2012;29:401-405. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 62] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 53. | Achen MG, Stacker SA. The vascular endothelial growth factor family; proteins which guide the development of the vasculature. Int J Exp Pathol. 1998;79:255-265. [PubMed] [Cited in This Article: ] |
| 54. | Gerwins P, Sköldenberg E, Claesson-Welsh L. Function of fibroblast growth factors and vascular endothelial growth factors and their receptors in angiogenesis. Crit Rev Oncol Hematol. 2000;34:185-194. [PubMed] [Cited in This Article: ] |
| 55. | Ferrara N. Role of vascular endothelial growth factor in the regulation of angiogenesis. Kidney Int. 1999;56:794-814. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 537] [Cited by in F6Publishing: 517] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 56. | Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9:669-676. [PubMed] [Cited in This Article: ] |
| 57. | Benjamin LE, Keshet E. Conditional switching of vascular endothelial growth factor (VEGF) expression in tumors: induction of endothelial cell shedding and regression of hemangioblastoma-like vessels by VEGF withdrawal. Proc Natl Acad Sci USA. 1997;94:8761-8766. [PubMed] [Cited in This Article: ] |
| 58. | Gerber HP, Dixit V, Ferrara N. Vascular endothelial growth factor induces expression of the antiapoptotic proteins Bcl-2 and A1 in vascular endothelial cells. J Biol Chem. 1998;273:13313-13316. [PubMed] [Cited in This Article: ] |
| 59. | Gerber HP, McMurtrey A, Kowalski J, Yan M, Keyt BA, Dixit V, Ferrara N. Vascular endothelial growth factor regulates endothelial cell survival through the phosphatidylinositol 3’-kinase/Akt signal transduction pathway. Requirement for Flk-1/KDR activation. J Biol Chem. 1998;273:30336-30343. [PubMed] [Cited in This Article: ] |
| 60. | Olsson AK, Dimberg A, Kreuger J, Claesson-Welsh L. VEGF receptor signalling - in control of vascular function. Nat Rev Mol Cell Biol. 2006;7:359-371. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2194] [Cited by in F6Publishing: 2273] [Article Influence: 126.3] [Reference Citation Analysis (0)] |
| 61. | Fox SB, Harris AL. Histological quantitation of tumour angiogenesis. APMIS. 2004;112:413-430. [PubMed] [Cited in This Article: ] |
| 62. | Iosifidou R, Galaktidou G, Ananiadis A, Bladika N, Patakiouta F, Bousoulegas A. VEGF-A, VEGF-C, VEGF-R2, EGFR and HER2 in serum plus EGFR in tissue of patients with triple-negative breast cancer. Breast Cancer Res [Internet]. 2009;11:P13. [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 63. | Chanana P, Pandey AK, Yadav BS, KaurJ , Singla S, DimriK , Trehan R, Krishan P. Significance of serum vascular endothelial growth factor and cancer antigen 15.3 in patients with triple negative breast cancer. JRP. 2014;13:60-67. [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 64. | Ali EM, Sheta M, Mohsen MA. Elevated serum and tissue VEGF associated with poor outcome in breast cancer patients. AJM. 2011;47:217–224. [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 65. | El-Arab LR, Swellam M, El Mahdy MM. Metronomic chemotherapy in metastatic breast cancer: impact on VEGF. J Egypt Natl Canc Inst. 2012;24:15-22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 66. | Taha FM, Zeeneldin AA, Helal AM, Gaber AA, Sallam YA, Ramadan H, Moneer MM. Prognostic value of serum vascular endothelial growth factor in Egyptian females with metastatic triple negative breast cancer. Clin Biochem. 2009;42:1420-1426. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 67. | Linderholm BK, Hellborg H, Johansson U, Elmberger G, Skoog L, Lehtiö J, Lewensohn R. Significantly higher levels of vascular endothelial growth factor (VEGF) and shorter survival times for patients with primary operable triple-negative breast cancer. Ann Oncol. 2009;20:1639-1646. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 214] [Cited by in F6Publishing: 235] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 68. | Andre F, Job B, Dessen P, Tordai A, Michiels S, Liedtke C, Richon C, Yan K, Wang B, Vassal G. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res. 2009;15:441-451. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 250] [Cited by in F6Publishing: 244] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 69. | Edling CE, Hallberg B. c-Kit--a hematopoietic cell essential receptor tyrosine kinase. Int J Biochem Cell Biol. 2007;39:1995-1998. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 182] [Cited by in F6Publishing: 178] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 70. | Schweizer J, Bowden PE, Coulombe PA, Langbein L, Lane EB, Magin TM, Maltais L, Omary MB, Parry DA, Rogers MA. New consensus nomenclature for mammalian keratins. J Cell Biol. 2006;174:169-174. [PubMed] [Cited in This Article: ] |
| 71. | Otterbach F, Bànkfalvi A, Bergner S, Decker T, Krech R, Boecker W. Cytokeratin 5/6 immunohistochemistry assists the differential diagnosis of atypical proliferations of the breast. Histopathology. 2000;37:232-240. [PubMed] [Cited in This Article: ] |
| 72. | Ross DT, Perou CM. A comparison of gene expression signatures from breast tumors and breast tissue derived cell lines. Dis Markers. 2001;17:99-109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 105] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 73. | Abd El-Rehim DM, Pinder SE, Paish CE, Bell J, Blamey RW, Robertson JF, Nicholson RI, Ellis IO. Expression of luminal and basal cytokeratins in human breast carcinoma. J Pathol. 2004;203:661-671. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 401] [Cited by in F6Publishing: 404] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 74. | van de Rijn M, Perou CM, Tibshirani R, Haas P, Kallioniemi O, Kononen J, Torhorst J, Sauter G, Zuber M, Köchli OR. Expression of cytokeratins 17 and 5 identifies a group of breast carcinomas with poor clinical outcome. Am J Pathol. 2002;161:1991-1996. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 408] [Cited by in F6Publishing: 436] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 75. | Thike AA, Iqbal J, Cheok PY, Chong AP, Tse GM, Tan B, Tan P, Wong NS, Tan PH. Triple negative breast cancer: outcome correlation with immunohistochemical detection of basal markers. Am J Surg Pathol. 2010;34:956-964. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 76. | Kim JM, Hwang TY, Kang SH, Lee SJ, Bae YK. Prognostic Significance of Basal Markers in Triple-negative Breast Cancers. J Breast Cancer. 2009;12:4-13. [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 77. | Rakha EA, Elsheikh SE, Aleskandarany MA, Habashi HO, Green AR, Powe DG, El-Sayed ME, Benhasouna A, Brunet JS, Akslen LA. Triple-negative breast cancer: distinguishing between basal and nonbasal subtypes. Clin Cancer Res. 2009;15:2302-2310. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 337] [Cited by in F6Publishing: 353] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 78. | Bryan BB, Schnitt SJ, Collins LC. Ductal carcinoma in situ with basal-like phenotype: a possible precursor to invasive basal-like breast cancer. Mod Pathol. 2006;19:617-621. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 159] [Cited by in F6Publishing: 171] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 79. | Kern SE, Kinzler KW, Bruskin A, Jarosz D, Friedman P, Prives C, Vogelstein B. Identification of p53 as a sequence-specific DNA-binding protein. Science. 1991;252:1708-1711. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 740] [Cited by in F6Publishing: 827] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 80. | Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307-310. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5100] [Cited by in F6Publishing: 4994] [Article Influence: 208.1] [Reference Citation Analysis (0)] |
| 81. | Karray-Chouayekh S, Baccouche S, Khabir A, Sellami-Boudawara T, Daoud J, Frikha M, Jlidi R, Gargouri A, Mokdad-Gargouri R. Prognostic significance of p16INK4a/p53 in Tunisian patients with breast carcinoma. Acta Histochem. 2011;113:508-513. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 82. | Alsner J, Yilmaz M, Guldberg P, Hansen LL, Overgaard J. Heterogeneity in the clinical phenotype of TP53 mutations in breast cancer patients. Clin Cancer Res. 2000;6:3923-3931. [PubMed] [Cited in This Article: ] |
| 83. | Kandioler-Eckersberger D, Ludwig C, Rudas M, Kappel S, Janschek E, Wenzel C, Schlagbauer-Wadl H, Mittlböck M, Gnant M, Steger G. TP53 mutation and p53 overexpression for prediction of response to neoadjuvant treatment in breast cancer patients. Clin Cancer Res. 2000;6:50-56. [PubMed] [Cited in This Article: ] |
| 84. | Hasebe T, Tamura N, Okada N, Hojo T, Akashi-Tanaka S, Shimizu C, Tsuda H, Shibata T, Sasajima Y, Iwasaki M. p53 expression in tumor-stromal fibroblasts is closely associated with the nodal metastasis and outcome of patients with invasive ductal carcinoma who received neoadjuvant therapy. Hum Pathol. 2010;41:262-270. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 85. | Dookeran KA, Dignam JJ, Ferrer K, Sekosan M, McCaskill-Stevens W, Gehlert S. p53 as a marker of prognosis in African-American women with breast cancer. Ann Surg Oncol. 2010;17:1398-1405. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 86. | Yamashita H, Nishio M, Toyama T, Sugiura H, Zhang Z, Kobayashi S, Iwase H. Coexistence of HER2 over-expression and p53 protein accumulation is a strong prognostic molecular marker in breast cancer. Breast Cancer Res. 2004;6:R24-R30. [PubMed] [Cited in This Article: ] |
| 87. | Linjawi A, Kontogiannea M, Halwani F, Edwardes M, Meterissian S. Prognostic significance of p53, bcl-2, and Bax expression in early breast cancer. J Am Coll Surg. 2004;198:83-90. [PubMed] [Cited in This Article: ] |
| 88. | Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, Pawitan Y, Hall P, Klaar S, Liu ET. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA. 2005;102:13550-13555. [PubMed] [Cited in This Article: ] |
| 89. | Biganzoli E, Coradini D, Ambrogi F, Garibaldi JM, Lisboa P, Soria D, Green AR, Pedriali M, Piantelli M, Querzoli P. p53 status identifies two subgroups of triple-negative breast cancers with distinct biological features. Jpn J Clin Oncol. 2011;41:172-179. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 90. | Chae BJ, Bae JS, Lee A, Park WC, Seo YJ, Song BJ, Kim JS, Jung SS. p53 as a specific prognostic factor in triple-negative breast cancer. Jpn J Clin Oncol. 2009;39:217-224. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 59] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 91. | Leong CO, Vidnovic N, DeYoung MP, Sgroi D, Ellisen LW. The p63/p73 network mediates chemosensitivity to cisplatin in a biologically defined subset of primary breast cancers. J Clin Invest. 2007;117:1370-1380. [PubMed] [Cited in This Article: ] |
| 92. | Burgess DJ, Doles J, Zender L, Xue W, Ma B, McCombie WR, Hannon GJ, Lowe SW, Hemann MT. Topoisomerase levels determine chemotherapy response in vitro and in vivo. Proc Natl Acad Sci USA. 2008;105:9053-9058. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 19] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 93. | Knoop AS, Knudsen H, Balslev E, Rasmussen BB, Overgaard J, Nielsen KV, Schonau A, Gunnarsdóttir K, Olsen KE, Mouridsen H. retrospective analysis of topoisomerase IIa amplifications and deletions as predictive markers in primary breast cancer patients randomly assigned to cyclophosphamide, methotrexate, and fluorouracil or cyclophosphamide, epirubicin, and fluorouracil: Danish Breast Cancer Cooperative Group. J Clin Oncol. 2005;23:7483-7490. [PubMed] [Cited in This Article: ] |
| 94. | Weigelt B, Horlings HM, Kreike B, Hayes MM, Hauptmann M, Wessels LF, de Jong D, Van de Vijver MJ, Van’t Veer LJ, Peterse JL. Refinement of breast cancer classification by molecular characterization of histological special types. J Pathol. 2008;216:141-150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 392] [Cited by in F6Publishing: 411] [Article Influence: 25.7] [Reference Citation Analysis (1)] |
| 95. | Urruticoechea A, Smith IE, Dowsett M. Proliferation marker Ki67 in early breast cancer. J Clin Oncol. 2005;23:7212-7220. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 546] [Cited by in F6Publishing: 573] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 96. | Zhou CJ, Zhang QH, Zhang TG, Sun SZ, Li H, Wang Y, Liu ZY. Expression of ER, Ki67 and cylinD1 in the pre-cancerous breast of Chinese patients. Pathol Oncol Res. 2009;15:153-158. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 97. | Harvey JA, Santen RJ, Petroni GR, Bovbjerg VE, Smolkin ME, Sheriff FS, Russo J. Histologic changes in the breast with menopausal hormone therapy use: correlation with breast density, estrogen receptor, progesterone receptor, and proliferation indices. Menopause. 2008;15:67-73. [PubMed] [Cited in This Article: ] |
| 98. | Fasching PA, Heusinger K, Haeberle L, Niklos M, Hein A, Bayer CM, Rauh C, Schulz-Wendtland R, Bani MR, Schrauder M. Ki67, chemotherapy response, and prognosis in breast cancer patients receiving neoadjuvant treatment. BMC Cancer. 2011;11:486. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 189] [Cited by in F6Publishing: 226] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 99. | Tanei T, Shimomura A, Shimazu K, Nakayama T, Kim SJ, Iwamoto T, Tamaki Y, Noguchi S. Prognostic significance of Ki67 index after neoadjuvant chemotherapy in breast cancer. Eur J Surg Oncol. 2011;37:155-161. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 100. | Selz J, Stevens D, Jouanneau L, Labib A, Le Scodan R. Prognostic value of molecular subtypes, ki67 expression and impact of postmastectomy radiation therapy in breast cancer patients with negative lymph nodes after mastectomy. Int J Radiat Oncol Biol Phys. 2012;84:1123-1132. [PubMed] [Cited in This Article: ] |
| 101. | de Azambuja E, Cardoso F, de Castro G, Colozza M, Mano MS, Durbecq V, Sotiriou C, Larsimont D, Piccart-Gebhart MJ, Paesmans M. Ki67 as prognostic marker in early breast cancer: a meta-analysis of published studies involving 12,155 patients. Br J Cancer. 2007;96:1504-1513. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 587] [Cited by in F6Publishing: 651] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 102. | Syed A, Giridhar PS, Sandhu K, Jader S, Al-Sam S, Sundaresan V. Ki67 in breast cancer patients and its correlation with clinico pathological factors. EJC. 2012;48:S121. [Cited in This Article: ] |
| 103. | Munzone E, Botteri E, Sciandivasci A, Curigliano G, Nole F, Rotmensz N. Prognostic significance of Ki67 in node negative(pN0), triple negative (TN) breast cancer(BC). 2011;General Poster Session, Breast Cancer-Triple-negative/Cytotoxics/Local Therapy; 2011 Jun 9; ASCO meeting abstracts, 2011: 1056. [Cited in This Article: ] |
| 104. | Keam B, Im SA, Lee KH, Han SW, Oh DY, Kim JH, Lee SH, Han W, Kim DW, Kim TY. Ki67 can be used for further classification of triple negative breast cancer into two subtypes with different response and prognosis. Breast Cancer Res. 2011;13:R22. [PubMed] [Cited in This Article: ] |
| 105. | Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O’Connor MJ. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123-134. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2792] [Cited by in F6Publishing: 2726] [Article Influence: 181.7] [Reference Citation Analysis (0)] |
| 106. | Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917-921. [PubMed] [Cited in This Article: ] |
| 107. | Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem. 2000;275:23899-23903. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 444] [Cited by in F6Publishing: 428] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 108. | Hastak K, Alli E, Ford JM. Synergistic chemosensitivity of triple-negative breast cancer cell lines to poly(ADP-Ribose) polymerase inhibition, gemcitabine, and cisplatin. Cancer Res. 2010;70:7970-7980. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 132] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 109. | Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913-917. [PubMed] [Cited in This Article: ] |
| 110. | Evers B, Drost R, Schut E, de Bruin M, van der Burg E, Derksen PW, Holstege H, Liu X, van Drunen E, Beverloo HB. Selective inhibition of BRCA2-deficient mammary tumor cell growth by AZD2281 and cisplatin. Clin Cancer Res. 2008;14:3916-3925. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 246] [Cited by in F6Publishing: 252] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 111. | Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci USA. 1994;91:8324-8328. [PubMed] [Cited in This Article: ] |
| 113. | Caldas-Lopes E, Cerchietti L, Ahn JH, Clement CC, Robles AI, Rodina A, Moulick K, Taldone T, Gozman A, Guo Y. Hsp90 inhibitor PU-H71, a multimodal inhibitor of malignancy, induces complete responses in triple-negative breast cancer models. Proc Natl Acad Sci USA. 2009;106:8368-8373. [PubMed] [Cited in This Article: ] |
| 114. | Liu CH, Chang SH, Narko K, Trifan OC, Wu MT, Smith E, Haudenschild C, Lane TF, Hla T. Overexpression of cyclooxygenase-2 is sufficient to induce tumorigenesis in transgenic mice. J Biol Chem. 2001;276:18563-18569. [PubMed] [Cited in This Article: ] |
| 115. | Half E, Tang XM, Gwyn K, Sahin A, Wathen K, Sinicrope FA. Cyclooxygenase-2 expression in human breast cancers and adjacent ductal carcinoma in situ. Cancer Res. 2002;62:1676-1681. [PubMed] [Cited in This Article: ] |
| 116. | Costa C, Soares R, Reis-Filho JS, Leitão D, Amendoeira I, Schmitt FC. Cyclo-oxygenase 2 expression is associated with angiogenesis and lymph node metastasis in human breast cancer. J Clin Pathol. 2002;55:429-434. [PubMed] [Cited in This Article: ] |
| 117. | Benoit V, Relic B, Leval Xd Xd, Chariot A, Merville MP, Bours V. Regulation of HER-2 oncogene expression by cyclooxygenase-2 and prostaglandin E2. Oncogene. 2004;23:1631-1635. [PubMed] [Cited in This Article: ] |
| 118. | Surowiak P, Materna V, Matkowski R, Szczuraszek K, Kornafel J, Wojnar A, Pudelko M, Dietel M, Denkert C, Zabel M. Relationship between the expression of cyclooxygenase 2 and MDR1/P-glycoprotein in invasive breast cancers and their prognostic significance. Breast Cancer Res. 2005;7:R862-R870. [PubMed] [Cited in This Article: ] |
| 119. | Hochgräfe F, Zhang L, O’Toole SA, Browne BC, Pinese M, Porta Cubas A, Lehrbach GM, Croucher DR, Rickwood D, Boulghourjian A. Tyrosine phosphorylation profiling reveals the signaling network characteristics of Basal breast cancer cells. Cancer Res. 2010;70:9391-9401. [PubMed] [Cited in This Article: ] |
| 120. | Finn RS, Dering J, Ginther C, Wilson CA, Glaspy P, Tchekmedyian N, Slamon DJ. Dasatinib, an orally active small molecule inhibitor of both the src and abl kinases, selectively inhibits growth of basal-type/”triple-negative” breast cancer cell lines growing in vitro. Breast Cancer Res Treat. 2007;105:319-326. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 301] [Cited by in F6Publishing: 305] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 121. | Faivre S, Kroemer G, Raymond E. Current development of mTOR inhibitors as anticancer agents. Nat Rev Drug Discov. 2006;5:671-688. [PubMed] [Cited in This Article: ] |
| 122. | Vinayak S, Carlson RW. mTOR inhibitors in the treatment of breast cancer. Oncology (Williston Park). 2013;27:38-44, 46, 48 passim. [PubMed] [Cited in This Article: ] |