Published online Aug 15, 2013. doi: 10.4291/wjgp.v4.i3.43
Revised: June 20, 2013
Accepted: July 4, 2013
Published online: August 15, 2013
Conventional triple therapies for Helicobacter pylori (H. pylori) eradication have recently shown a disappointing reduction in effectiveness in many countries. The main reason for failure was found to be bacterial resistance to one of the most commonly used antibiotics, clarithromycin. An additional problem for conventional triple therapy is the high rate of resistance to metronidazole found in Europe, America and Asia. In Italy, in the last 15 years a 2-fold increase in resistance has occurred. A recent study of the whole of Italy included about 20 patients from each region at the first endoscopic diagnosis of H. pylori infection. The most surprising result was the patchy distribution of resistance, which was almost absent in two regions (one northern and one southern), although the highest prevalence was found in some regions of the South. In the paediatric population we found a 25% prevalence of resistance in a sample of H. pylori positive children observed between 2002 and 2007, mirroring data obtained in southern European countries. Clarithromycin resistance assessment is currently based on phenotypic detection performed after culture the agar dilution method or E-test, and genotypic methods based on polymerase chain reaction (PCR). In a recent comparative study we found a 71.2% agreement between the two methods. Culture-free techniques are highly accurate in finding even minimal traces of genotypically resistant strains. Moreover, PCR-based tools are accurate in detecting a heteroresistant status, defined as the co-existence of some strains that are susceptible and some resistant to the same antibiotic in an individual patient. Three point mutations, namely A2143G, A2142G and A2142C, are responsible for 90% of cases of primary clarithromycin resistance in H. pylori strains isolated in Western countries, although we previously demonstrated that the presence of the A2143G mutation, but not A2142G or A2142C, significantly lowered the H. pylori eradication rate. Treatment failure has considerable cost/benefit implications because of “waste” of National Health System and patient resources, in terms of drugs, further diagnostic tests and medical examination expenses. Therefore, in future it would be very useful to be able to test for clarithromycin resistance before starting conventional triple therapy. Hopefully, fast, effective non-invasive tests may soon be devised to determine this condition.
Core tip: Clarithromycin resistance is the main reason for the failure of conventional therapies for Helicobacter pylori. We evaluate the scope of the problem, as reported in literature and especially on the basis of personal data in adult and paediatric populations. Another issue is the detection of resistance using phenotypic and genotypic methods; comparison is made of the limits and advantages of these approaches. Cost/benefit analysis of unsuccessful eradication therapy is performed. Based on these considerations, the best solution in future seems likely to be the detection of resistant strains before starting treatment.
-
Citation: Giorgio F, Principi M, De Francesco V, Zullo A, Losurdo G, Di Leo A, Ierardi E. Primary clarithromycin resistance to
Helicobacter pylori : Is this the main reason for triple therapy failure? World J Gastrointest Pathophysiol 2013; 4(3): 43-46 - URL: https://www.wjgnet.com/2150-5330/full/v4/i3/43.htm
- DOI: https://dx.doi.org/10.4291/wjgp.v4.i3.43
Helicobacter pylori (H. pylori) eradication may have clinical implications with a very high impact: healing of peptic ulcer disease, resolution of low-grade lymphoma in about 80% of cases and the prevention of gastric adenocarcinoma[1,2]. However, some problems have arisen in clinical practice in recent years, namely the decreased eradication rate of conventional therapies and the onset and diffusion of antibiotic resistances worldwide[3]. Triple therapy, comprising treatment with two antibiotics, amoxicillin and clarithromycin, and a proton pump inhibitor for a week, was recommended as the treatment of choice at several consensus conferences. However, this treatment has recently shown a disappointing reduction in effectiveness in many countries and the main reason for failure has been found to be bacterial resistance to one of the most commonly used antibiotics, clarithromycin[4].
An assessment of primary clarithromycin resistance made about 10 years ago demonstrated differences between children and adults in Europe. In adults, a marked difference has been observed between Northern countries, where the overall prevalence was about 5%, and Southern countries, where the resistance rate was to up to 20%. This difference was not observed in children. Indeed, in all European countries a virtually equal, high prevalence has been reported, ranging from 12.5% to 23.5%. An additional problem for conventional triple therapy is the high rate of resistance to metronidazole[5,6] found in Europe (from 23.5% in Spain to 40.3% in the United Kingdom), America (from 21.6% in the United States to 76.3% in Mexico) and Asia (from 9%-12% in Japan to 41.9% in Korea).
In Italy, data obtained 10 years ago showed a strong difference between the resistance rates in Northern (2%) and Central (20%) areas[7,8]. However, in the same period a study by our group found a median value of 28.7% in two areas in Southern and Central Italy[9]. In 2007, in a retrospective study performed over a 15-year period in the same geographical area, we found a 10.2% prevalence of primary clarithromycin resistance in 147 patients in the 1990 period, whereas by 2004-2005 the resistance found in 178 patients had increased significantly, to 21.3% or 2-fold higher[10]. The phenomenon was found to be more evident in females. A possible explanation for this finding may be a wider use of the antibiotic in the course of infections with a greater prevalence in the female sex, such as urinary tract infections[11,12].
A more recent study (2011) was focused on the whole of Italy, including about 20 patients from each region at the first endoscopic diagnosis of H. pylori infection. The most surprising result was the patchy distribution of resistance, which was almost absent in two regions (one Northern and one Southern) although the highest prevalence was found in some regions of the South (20%-25%). Due to this distribution variability, the overall resistance rate was lower than expected (about 10%). Resistances were higher in patients with non-ulcer dyspepsia than in those with peptic ulcer, in agreement with findings in previous investigations[13].
In the paediatric population we found a 25% prevalence of resistance in a sample of 168 H. pylori positive children observed between 2002 and 2007, mirroring other data from European countries, where high values reflect the environmental conditions of southern areas[14].
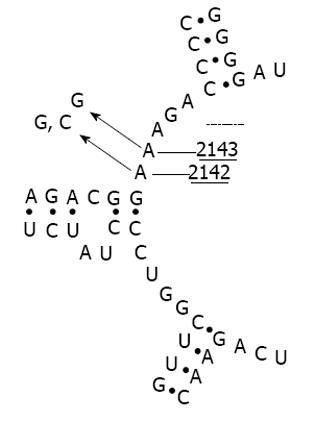
Clarithromycin resistance assessment is currently based on phenotypic detection performed after culture and the agar dilution method or E-test. However, in the past decade, different polymerase chain reaction (PCR) based approaches have been developed as alternative tools[15]. These techniques allow assessment of mutations in the peptidyltransferase region encoded in domain V of the H. pylori 23S ribosomal RNA region that confers clarithromycin resistance[16,17]. Undeniably, both culture and PCR-based methods have both advantages and limitations[18]. Bacterial culture allows an overall evaluation of H. pylori clarithromycin resistance, regardless of the intrinsic mechanism involved (point mutations, RNA methylations, efflux pumps, etc.). Nevertheless, H. pylori is a troublesome bacterium and culture may be difficult even in expert hands. Indeed, sensitivity values of culture as low as 55%-73% have been reported in some trials[19]. By contrast, PCR-based culture-free techniques are highly accurate in finding even minimal traces of genotypically resistant strains. Moreover, PCR-based tools are accurate in detecting a heteroresistant status, defined as the co-existence of some strains that are susceptible and some resistant to the same antibiotic in an individual patient. These techniques may be used even on paraffin-embedded tissue[20]. Nevertheless, these approaches are unable to detect clarithromycin resistance when it is based on uncommon genetic mechanisms (deletions, RNA methylations, etc.). Although several mutations have been detected, it has been found that three point mutations, namely A2143G, A2142G and A2142C (Figure 1), are responsible for 90% of cases of primary clarithromycin resistance in H. pylori strains isolated in Western countries[21,22]. Some studies showed that these point mutations are associated with different MIC values for clarithromycin resistance assessed by culture in vitro[23], suggesting that they might have a different impact on the therapeutic outcome. Indeed, we previously found that the presence of the A2143G mutation, but not A2142G or A2142C, significantly lowered the H. pylori eradication rate[24,25].
In a recent comparative study we found that the prevalence of clarithromycin phenotypic resistance was significantly lower than that of genotypic resistance (18.4% vs 37.6%, P < 0.001). An agreement between the two methods was present in 71.2% of cases. A significant difference in the eradication rate was seen between clarithromycin-susceptible and resistant strains, when assessed with either the E test (92.4% vs 55.5%, P < 0.001) or a PCR-based method (94.5% vs 70.9%, P < 0.001). Notably, the eradication rate showed the lowest value (30.7%) when the phenotypic bacterial resistance was genetically linked to the A2143G point mutation. The conclusion of this study was that there is a disagreement between the two methods; clarithromycin resistance markedly reduces H. pylori eradication only when it is linked to a specific point mutation[26].
On these grounds, it has been suggested that MIC values for clarithromycin resistance should be dropped to 0.5 in order to improve the efficacy of phenotypic detection[27].
Conventional 7-d triple therapy failure leads to a “waste” of about 27 € in each case of resistance to clarithromycin. The sum is doubled when the therapy duration is prolonged to 10-14 d, as suggested by some reports[28]. If sequential therapy is used, the cost of treatment is about 40 € but the therapeutic gain is unsatisfactory.
In addition to the net cost of drugs, the cost of support treatment used in cases of side effects (probiotics, prokinetics, anti-acids), accounts for a further 5-25 € on average. In cases of therapeutic failure the patient will undergo a new medical examination (average cost: 36 €) and new surveys for H. pylori status by non-invasive (urea breath test: 70 €, stool antigen: 30-40 €) and invasive methods (esophagogastroduodenoscopy: 220 + 36 € of individual contribution; biopsy: 40 €; histological examination: 95 + 36 € of individual contribution)[29].
In this scenario it is clear that inappropriate use of an anti H. pylori regimen incurs heavy costs not only in terms of the initial, ineffective treatment but also of the later additional visits and investigations.
In future it will be more effective to test for clarithromycin resistance before starting conventional triple therapy. The limit of this approach is the high cost of molecular analysis, as well as the difficulty of performing phenotypic investigations, even for experts. Other issues hindering a wider spread of these assessments in clinical practice are the long time between testing and obtaining the results on which to plan therapy, and the need to perform an invasive examination (esophagogastro-duodenoscopy). In conclusion, it would be very useful to be able to test for clarithromycin resistance before starting conventional triple therapy. Hopefully, fast, effective non-invasive tests may soon be devised to determine this condition.
P- Reviewers Figura N, Schwarz SM S- Editor Gou SX L- Editor Hughes D E- Editor Li JY
| 1. | Graham DY, Lew GM, Klein PD, Evans DG, Evans DJ, Saeed ZA, Malaty HM. Effect of treatment of Helicobacter -pylori infection on the long-term recurrence of gastric or duodenal ulcer. A randomized, controlled study. Ann Intern Med. 1992;116:705-708. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 672] [Cited by in F6Publishing: 611] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 2. | Malfertheiner P, Mégraud F, O’Morain C, Hungin AP, Jones R, Axon A, Graham DY, Tytgat G. Current concepts in the management of Helicobacter pylori infection--the Maastricht 2-2000 Consensus Report. Aliment Pharmacol Ther. 2002;16:167-180. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 846] [Cited by in F6Publishing: 830] [Article Influence: 37.7] [Reference Citation Analysis (1)] |
| 3. | De Francesco V, Giorgio F, Hassan C, Manes G, Vannella L, Panella C, Ierardi E, Zullo A. Worldwide H. pylori antibiotic resistance: a systematic review. J Gastrointestin Liver Dis. 2010;19:409-414. [PubMed] [Cited in This Article: ] |
| 4. | Mégraud F. H pylori antibiotic resistance: prevalence, importance, and advances in testing. Gut. 2004;53:1374-1384. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 607] [Cited by in F6Publishing: 653] [Article Influence: 32.7] [Reference Citation Analysis (1)] |
| 5. | Perez Aldana L, Kato M, Nakagawa S, Kawarasaki M, Nagasako T, Mizushima T, Oda H, Kodaira J, Shimizu Y, Komatsu Y. The relationship between consumption of antimicrobial agents and the prevalence of primary Helicobacter pylori resistance. Helicobacter. 2002;7:306-309. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 99] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 6. | Kato M, Yamaoka Y, Kim JJ, Reddy R, Asaka M, Kashima K, Osato MS, El-Zaatari FA, Graham DY, Kwon DH. Regional differences in metronidazole resistance and increasing clarithromycin resistance among Helicobacter pylori isolates from Japan. Antimicrob Agents Chemother. 2000;44:2214-2216. [PubMed] [Cited in This Article: ] |
| 7. | Toracchio S, Marzio L. Primary and secondary antibiotic resistance of Helicobacter pylori strains isolated in central Italy during the years 1998-2002. Dig Liver Dis. 2003;35:541-545. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 63] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 8. | Pilotto A, Rassu M, Leandro G, Franceschi M, Di Mario F. Prevalence of Helicobacter pylori resistance to antibiotics in Northeast Italy: a multicentre study. GISU. Interdisciplinary Group for the Study of Ulcer. Dig Liver Dis. 2000;32:763-768. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 66] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 9. | de Francesco V, Margiotta M, Zullo A, Hassan C, Valle ND, Burattini O, Cea U, Stoppino G, Amoruso A, Stella F. Primary clarithromycin resistance in Italy assessed on Helicobacter pylori DNA sequences by TaqMan real-time polymerase chain reaction. Aliment Pharmacol Ther. 2006;23:429-435. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 36] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 10. | De Francesco V, Margiotta M, Zullo A, Hassan C, Giorgio F, Burattini O, Stoppino G, Cea U, Pace A, Zotti M. Prevalence of primary clarithromycin resistance in Helicobacter pylori strains over a 15 year period in Italy. J Antimicrob Chemother. 2007;59:783-785. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 77] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 11. | August SL, De Rosa MJ. Evaluation of the prevalence of urinary tract infection in rural Panamanian women. PLoS One. 2012;7:e47752. [PubMed] [Cited in This Article: ] |
| 12. | Boyanova L, Ilieva J, Gergova G, Davidkov L, Spassova Z, Kamburov V, Katsarov N, Mitov I. Numerous risk factors for Helicobacter pylori antibiotic resistance revealed by extended anamnesis: a Bulgarian study. J Med Microbiol. 2012;61:85-93. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 13. | De Francesco V, Giorgio F, Ierardi E, Zotti M, Neri M, Milano A, Varasano V, Luzza F, Suraci E, Marmo R. Primary clarithromycin resistance in Helicobacter pylori: the Multicentric Italian Clarithromycin Resistance Observational (MICRO) study. J Gastrointestin Liver Dis. 2011;20:235-239. [PubMed] [Cited in This Article: ] |
| 14. | Francavilla R, Lionetti E, Castellaneta S, Margiotta M, Piscitelli D, Polimeno L, Cavallo L, Ierardi E. Clarithromycin-Resistant Genotypes and Eradication of Helicobacter Pylori. J Pediatr. 2010;157:228-232. [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 15. | Francesco VD, Zullo A, Hassan C, Giorgio F, Rosania R, Ierardi E. Mechanisms of Helicobacter pylori antibiotic resistance: An updated appraisal. World J Gastrointest Pathophysiol. 2011;2:35-41. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 92] [Cited by in F6Publishing: 91] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 16. | Wada T, Maeda S, Tamaru A, Imai S, Hase A, Kobayashi K. Dual-probe assay for rapid detection of drug-resistant Mycobacterium tuberculosis by real-time PCR. J Clin Microbiol. 2004;42:5277-5285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 58] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 17. | Baglan PH, Bozdayi G, Ozkan M, Ahmed K, Bozdayi AM, Ozden A. Clarithromycin resistance prevalence and Icea gene status in Helicobacter Pylori clinical isolates in Turkish patients with duodenal ulcer and functional dyspepsia. J Microbiol. 2006;44:409-416. [PubMed] [Cited in This Article: ] |
| 18. | Monno R, Giorgio F, Carmine P, Soleo L, Cinquepalmi V, Ierardi E. Helicobacter pylori clarithromycin resistance detected by Etest and TaqMan real-time polymerase chain reaction: a comparative study. APMIS. 2012;120:712-717. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 19. | Zullo A, Hassan C, Lorenzetti R, Winn S, Morini S. A clinical practice viewpoint: to culture or not to culture Helicobacter pylori? Dig Liver Dis. 2003;35:357-361. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 64] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 20. | Paireder S, Werner B, Bailer J, Werther W, Schmid E, Patzak B, Cichna-Markl M. Comparison of protocols for DNA extraction from long-term preserved formalin fixed tissues. Anal Biochem. 2013;439:152-160. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 21. | Matsumura M, Hikiba Y, Ogura K, Togo G, Tsukuda I, Ushikawa K, Shiratori Y, Omata M. Rapid detection of mutations in the 23S rRNA gene of Helicobacter pylori that confers resistance to clarithromycin treatment to the bacterium. J Clin Microbiol. 2001;39:691-695. [PubMed] [Cited in This Article: ] |
| 22. | Oleastro M, Ménard A, Santos A, Lamouliatte H, Monteiro L, Barthélémy P, Mégraud F. Real-time PCR assay for rapid and accurate detection of point mutations conferring resistance to clarithromycin in Helicobacter pylori. J Clin Microbiol. 2003;41:397-402. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 155] [Cited by in F6Publishing: 152] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 23. | De Francesco V, Zullo A, Ierardi E, Vaira D. Minimal inhibitory concentration (MIC) values and different point mutations in the 23S rRNA gene for clarithromycin resistance in Helicobacter pylori. Dig Liver Dis. 2009;41:610-611. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 22] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 24. | De Francesco V, Margiotta M, Zullo A, Hassan C, Troiani L, Burattini O, Stella F, Di Leo A, Russo F, Marangi S. Clarithromycin-resistant genotypes and eradication of Helicobacter pylori. Ann Intern Med. 2006;144:94-100. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 138] [Cited by in F6Publishing: 147] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 25. | De Francesco V, Zullo A, Ierardi E, Giorgio F, Perna F, Hassan C, Panella C, Vaira D. The A2143G point mutation of clarithromycin resistance affects Helicobacter pylori eradication. J Clin Gastroenterol. 2009;43:386. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 26. | De Francesco V, Zullo A, Ierardi E, Giorgio F, Perna F, Hassan C, Morini S, Panella C, Vaira D. Phenotypic and genotypic Helicobacter pylori clarithromycin resistance and therapeutic outcome: benefits and limits. J Antimicrob Chemother. 2010;65:327-332. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 77] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 27. | European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 2.0, valid from 2012-01-01: 67. Available from: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/EUCAST_breakpoints_v_2.0_120101.pdf. [Cited in This Article: ] |
| 28. | De Francesco V, Della Valle N, Stoppino V, Amoruso A, Muscatiello N, Panella C, Ierardi E. Effectiveness and pharmaceutical cost of sequential treatment for Helicobacter pylori in patients with non-ulcer dyspepsia. Aliment Pharmacol Ther. 2004;19:993-998. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 29. | Italian pharmaceutical reference book 2013. Health Care Network. Euromedia s. r. l. Available from: http://www.prontuariofarmaceutico.it/. [Cited in This Article: ] |