- 1Division of Infection and Immunity, Walter and Eliza Hall Institute of Medical Research, Parkville, VIC, Australia
- 2Department of Medical Biology, The University of Melbourne, Parkville, VIC, Australia
After more than 50 years of intensive research and development, only one malaria vaccine candidate, “RTS,S,” has progressed to Phase 3 clinical trials. Despite only partial efficacy, this candidate is now forecast to become the first licensed malaria vaccine. Hence, more efficacious second-generation malaria vaccines that can significantly reduce transmission are urgently needed. This review will focus on a major obstacle hindering development of effective malaria vaccines: parasite antigenic diversity. Despite extensive genetic diversity in leading candidate antigens, vaccines have been and continue to be formulated using recombinant antigens representing only one or two strains. These vaccine strains represent only a small fraction of the diversity circulating in natural parasite populations, leading to escape of non-vaccine strains and challenging investigators’ abilities to measure strain-specific efficacy in vaccine trials. Novel strategies are needed to overcome antigenic diversity in order for vaccine development to succeed. Many studies have now cataloged the global diversity of leading Plasmodium falciparum and Plasmodium vivax vaccine antigens. In this review, we describe how population genetic approaches can be applied to this rich data source to predict the alleles that best represent antigenic diversity, polymorphisms that contribute to it, and to identify key polymorphisms associated with antigenic escape. We also suggest an approach to summarize the known global diversity of a given antigen to predict antigenic diversity, how to select variants that best represent the strains circulating in natural parasite populations and how to investigate the strain-specific efficacy of vaccine trials. Use of these strategies in the design and monitoring of vaccine trials will not only shed light on the contribution of genetic diversity to the antigenic diversity of malaria, but will also maximize the potential of future malaria vaccine candidates.
Introduction
Malaria is the most devastating parasitic disease afflicting humankind. The disease results from infection with protozoan parasites of the genus, Plasmodium and is transmitted by female anophelene mosquitoes. Of the 3.4 billion people in 108 countries at risk of malaria, 1.2 billion are at high risk of disease. In 2012, it was estimated that this disease caused 2000 deaths per day, the majority (77%) being children <5 years of age in sub-Saharan Africa infected with Plasmodium falciparum, the most virulent of the five known human malaria parasites (1, 2). In addition to this enormous heath toll, malaria exerts a heavy economic burden contributing to the cycle of poverty in many resource-limited settings (3). Although less lethal than P. falciparum, the majority of malaria infections occurring outside of sub-Saharan Africa are caused by Plasmodium vivax, with as many as 2.3 billion people at risk of infection (4). Several unique features of P. vivax biology, including its dormant stage in the human liver, make it more resistant to malaria elimination. As a result, P. vivax is predicted to present the ultimate obstacle to malaria elimination in endemic countries (5). Nevertheless, research into this parasite lags far behind that of P. falciparum due to its relatively recent recognition as a serious threat to global public health and lack of a viable long term in vitro culture system (4, 6).
Intensified malaria control efforts, supported by the Roll Back Malaria campaign, have resulted in a 42% decrease in malaria deaths worldwide in the last decade and many previously endemic countries have now shifted from controlling malaria to an elimination agenda (1). In 2007, encouraged by the stunning impact of this campaign, major funding bodies united to issue the ultimate challenge, to eradicate malaria globally by progressive malaria elimination from different countries and regions (3, 7). From past malaria eradication attempts, it is clear that in order for this ambitious goal to be achieved, malaria transmission must be permanently interrupted. Interventions that reduce the parasite reservoir, limit the rate at which infections are spread and the duration of time that a human or mosquito host is infectious are therefore urgently needed (8). In concert with other malaria control interventions, this could be achieved with the development of a broadly effective malaria vaccine.
Malaria parasites are ancient organisms with abundant genetic polymorphisms, much of which have evolved to escape host immune responses and thus presents a major obstacle to the development of a vaccine that provides broad protection against all, or at least the majority of strains (9). As with other pathogens, the challenge in developing an effective malaria vaccine will be to differentiate between diversity that is associated with immune escape and cross protection, and that which has no bearing on the immune response, having simply accumulated over time through genetic drift or through adaptation to diverse host environments (9). To date, the polymorphisms in malaria antigens targeted by functionally important antibodies remain poorly characterized (10). Very little is known of how sequence polymorphisms relate to antigenic diversity or the potential for polymorphisms to mediate vaccine escape for Plasmodium spp. (11). The key to success with other pathogens has been the identification of immunologically relevant diversity. This has been achieved by performing population genetic and structural studies to identify functionally relevant polymorphisms, followed by molecular epidemiological surveys or in vitro functional studies prior to development and testing of vaccines (9). Narrowing the focus to immunologically relevant polymorphisms would greatly reduce the diversity that must be considered when developing multivalent malaria vaccines covering a broad range of strains (2, 9, 12) (Box 1).
Box 1. Glossary of terms.
Strain: a parasite variant that is genetically unique and induces specific immune responses against one or more of its antigens.
Isolate: a parasite specimen derived from an infected individual that has been either adapted for in vitro culture or used directly for experiments. Different isolates from the same population may contain parasites that are genetically identical at one or more loci. Individual isolates may also contain one or more genetically distinct parasites if they have been collected from an individual with multiple infections.
Clone: a genetically homogeneous parasite isolate.
Polymorphism: variation in the population at a particular nucleotide or amino acid residue.
Allele: one variant of a particular genetic locus. It can refer to individual polymorphic sites within a nucleotide or amino acid sequence, or the combination of all polymorphic sites in a gene or gene region (known as the haplotype).
Haplotype: a combination of alleles in a gene or gene region carried by a particular parasite clone.
Serotype: a haplotype from a gene or gene region that is antigenically unique and induces strain-specific responses.
Monovalent vaccine: a vaccine containing only one distinct antigen or one allele of the same antigen.
Multivalent vaccine: a vaccine containing two or more distinct antigens, or two or more alleles of the same antigen.
Malaria Vaccines: Past, Present, and Future
A long lasting, broadly efficacious malaria vaccine would be the most sustainable approach to control and eventually eradicate malaria. That a malaria vaccine may be feasible is strongly supported by the fact that people living in malaria endemic areas develop protective immunity against malaria symptoms during childhood (13). By adulthood, decreases in the prevalence of infection and density of parasitemia are achieved indicating that this immunity eventually provides some protection against infection (14, 15). Passive transfer of immunoglobulin from hyper-immune African adults to non-immune children with severe malaria was shown to have curative properties, demonstrating that antibody responses are largely responsible for protection against clinical disease (16). Furthermore, vaccination of rodent and primate models with recombinant parasite antigens elicits high antibody titers that are associated with protection against subsequent malaria challenge (17–20). Although the development of malaria vaccines has been an active focus of the malaria research community over the last 50 years, a vaccine remains a missing component of malaria control and elimination strategies (21).
Despite numerous promising malaria vaccine candidates and several partially successful malaria vaccine trials (22–25), short-lived protection, limited funding, and a lack of key technologies has hampered further testing and the scale-up of clinical trials of novel malaria vaccine candidates. The first successful malaria vaccine trial, based on a “whole parasite” approach, was conducted in humans in the 1950s. Vaccination with irradiated sporozoites was shown to protect against both homologous and heterologous challenge in humans (26, 27). However, the need for large-scale production prevented further development of this approach. With the advent of molecular technologies in the 1980s, the focus shifted to so-called “subunit” approaches, including highly immunogenic parasite antigens as targets such as the circumsporozoite protein (CSP), which was identified as the parasite antigenic determinant targeted by immune responses induced by the sporozoite vaccine (28). Following these early studies, and on the back of highly promising pre-clinical studies, many small-scale subunit vaccine trials were conducted in humans. Efficacy was highly variable with many candidates demonstrating no protective effect, however, there were some promising candidates identified that continue to be further developed today (21), and these are discussed in more detail below.
In the last decade, there has been an attempt to assist and accelerate development of a malaria vaccine with the establishment of the PATH Malaria Vaccine Initiative (MVI), which has greatly progressed the evaluation and identification of promising malaria vaccine candidates. To further focus and unite the global vaccine effort, in 2006 the World Health Organization (WHO) launched the first malaria vaccine technology roadmap with the landmark goal:
By 2015, develop and license a first generation malaria vaccine that has a protective efficacy of more than 50% against severe disease and death and lasts more than one year (29).
Currently, a single vaccine candidate is on track to meet this goal. Known as “RTS,S,” this vaccine is based on the repeat region and T-cell epitopes of CSP. RTS,S is the subject of a large multicentre Phase 3 trial involving more than 15,000 children over 11 sites in sub-Saharan Africa. The full results of the trial have not yet been published; however, after three doses, clinical malaria cases during the first 18 months of follow-up decreased by an estimated 46% (severe disease by 36%) in children 5–17 months of age at first vaccination, and 27% (severe disease by 15%) in infants aged 6–12 weeks of age at first vaccination (30), therefore, RTS,S efficacy is approaching the above-mentioned WHO criteria for a first generation malaria vaccine. As a result, this vaccine is likely to be licensed in 2015 for use in young African children and could lead to significant decreases in malaria morbidity and mortality in this high-risk population. Despite this positive outlook, it is cautioned that this vaccine is only partially protective against disease and wanes over time. New, second-generation vaccines will need to have major improvements in efficacy to meet the challenges ahead (10).
Recognizing the changing epidemiology of malaria in the context of a shrinking global malaria map and a shift in populations most at risk of infection (5), as well as the need for vaccines with higher efficacies than RTS,S if malaria elimination is to be achieved, the goals of the Malaria Vaccine Technology Roadmap were recently reset with two major objectives:
By 2030, license vaccines targeting P. falciparum and P. vivax that encompass: (i) development of second-generation malaria vaccines that provide a protective efficacy of more than 75% against clinical (mild and severe) malaria … and (ii) development of malaria vaccines that reduce transmission of the parasite and thereby reduce the incidence of human infection … (31).
To achieve these new goals, a better understanding of the minimal requirements and mechanisms underlying development of immunity against P. falciparum and P. vivax (10, 12) as well as knowledge regarding the factors that influence transmission of both species, will be essential. As the development of vaccines against P. vivax is also a major goal, it will be important to consider the distinct features of this species, which underscores the need to intensify research efforts into this relatively neglected parasite.
Approaches to Malaria Vaccine Development
Malaria parasites are complex eukaryotes comprised of many antigenic targets. It has been suggested that vaccines may need to be as complex as the parasite itself (32) and therefore there has been considerable interest in the whole parasite approach. As mentioned above, irradiated live sporozoite vaccinations have shown great success in clinical trials (27, 33, 34). Currently, different methods are being used to attenuate sporozoite stages including chemical and genetic modification, and these have been reviewed elsewhere (35). However, there are some challenges to overcome in addition to the technical difficulties and high costs associated with scaling up production including the dose required to induce long-lasting protective immunity, transport, and storage in the absence of a reliable cold chain for distribution to at risk populations (36). There is also a risk of reversion to virulence (12). Hence the alternative subunit vaccine approach continues to be vigorously evaluated (37). As indicated above, this involves individual recombinant parasite proteins administered as monovalent preparations or combinations of multiple proteins together with different vectors and adjuvants that enhance the immune response. As the majority of clinical trials conducted to date have been based on subunit vaccines, this rest of this review will focus on the development of this class of malaria vaccines and the challenges associated with this approach.
Several highly abundant parasite proteins were identified as targets of natural immunity many years ago but in recent years the list of possible candidates has expanded. These candidates have been extensively validated in pre-clinical studies using different approaches including the measurement of inhibitory antibody responses in in vitro growth and invasion assays (short term culture only for P. vivax) (18, 38–41) and by vaccinating animal models followed by challenge infections (17, 18, 42–45). Subunit vaccines have been or are being developed based on these advanced candidate antigens, which are expressed in almost every stage of the parasite lifecycle. They have been classified into one of three different groups based on the target lifecycle stage (Figure 1) (46):
(i) pre-erythrocytic vaccines: these vaccines aim to prevent infection by targeting the infective stage, known as the sporozoite (e.g., RTS,S). Alternatively, pre-erythrocytic vaccines can target antigens expressed by liver stage parasites to prevent the emergence of merozoites into the bloodstream, the stage of infection responsible for the clinical symptoms of malaria infection (Figure 1). The risk associated with targeting sporozoite antigens is that the antigen dose inoculated during a natural infection is very low, with only a small number of sporozoites injected by the vector (~20), and this may not be sufficient to elicit an effective host immune response. Additionally, only one sporozoite needs to escape the vaccine-mediated immune response and invade liver cells for ~10,000 infectious merozoites to be produced, resulting in blood stage infection and clinical disease (47).
(ii) blood stage vaccines: the vast majority of malaria vaccine candidates are designed to protect against the blood stage of infection (Figure 1). Since all of the symptoms of malaria occur during this stage, the majority of vaccines targeting antigens expressed during the blood stage are designed primarily to prevent disease. One approach is to target merozoite antigens to prevent red blood cell invasion and reduce the density and prevalence of parasites in the infected host (Figure 1). In principle, this reduction in parasite density may also reduce the density of transmission forms, known as gametocytes (i.e., the sexual stage of the parasite transmitted from human to mosquito host) (Figure 1). In addition to preventing clinical disease, an effective blood stage vaccine that reduces parasite density may therefore also contribute to reducing malaria transmission (10, 46, 48). Approaches are also being developed to target the major surface protein expressed on the P. falciparum infected red blood cell known as erythrocyte membrane protein 1 (PfEMP1). PfEMP1 mediates adhesion to host cells, a mechanism that is associated with severe malaria [reviewed by Hviid (49)].
(iii) transmission-blocking vaccines: the aim of transmission-blocking vaccines is to target antigens expressed during lifecycle stages in the mosquito host (e.g., gametocyte or oocyst antigens) (Figure 1). Although these vaccines would not directly prevent infection or clinical disease, they would greatly assist elimination efforts to prevent the onward transmission of infections that may be imported into an elimination zone (10, 46).
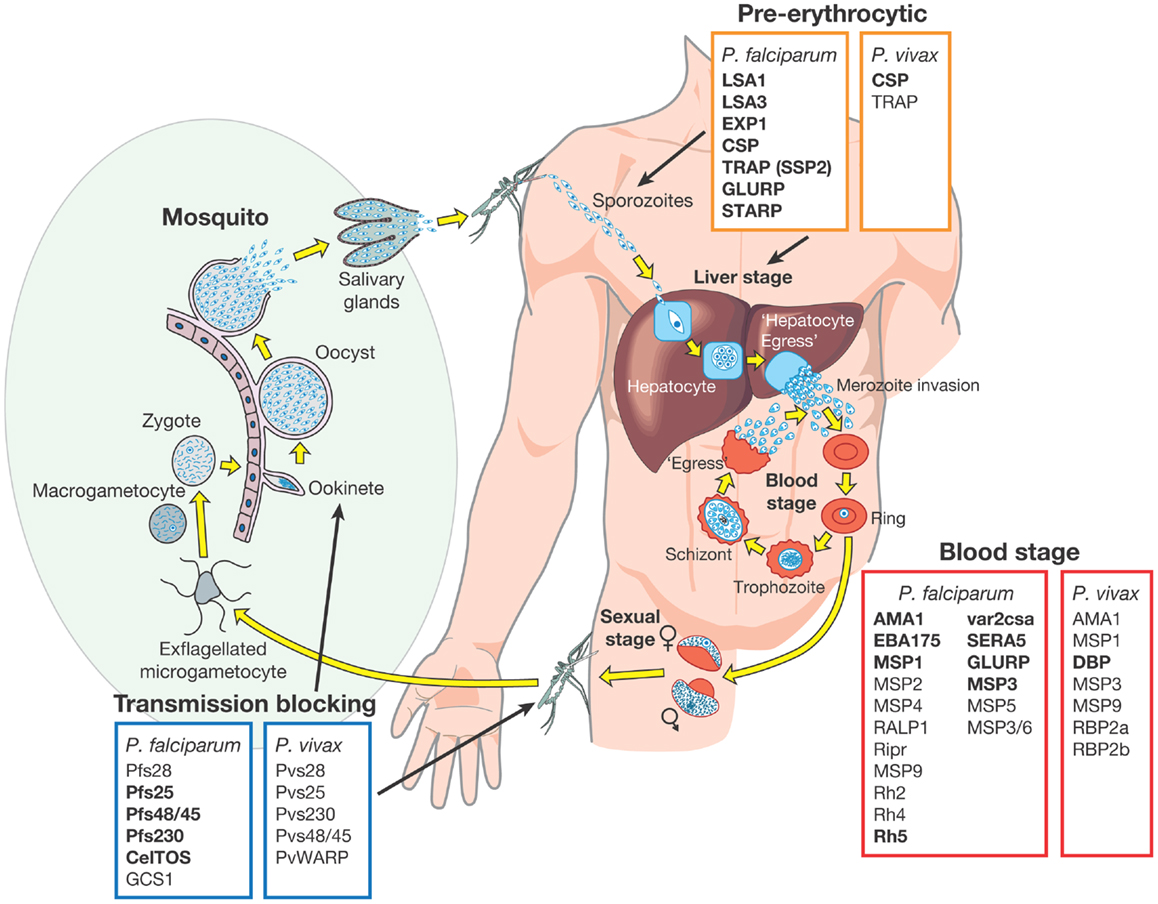
Figure 1. Malaria vaccine candidate antigens are shown. All candidate antigens for Plasmodium falciparum and Plasmodium vivax are superimposed on the Plasmodium lifecycle, to indicate the category of malaria vaccine being developed and the lifecycle stage targeted. Antigens indicated in bold are those that are currently being evaluated in pre-clinical trials or have entered at least Phase 1 clinical trials according to the WHO malaria vaccine rainbow tables (50). The P. vivax latent stages known as “hyponozoites” are not shown but these occur in the liver stage.
Development of a vaccine against P. falciparum is well advanced with 31 promising antigens identified (Figure 1). Currently, 27 subunit candidates comprising different domains and alleles for 22 different antigens are being tested in pre-clinical or clinical trials (50) (Figure 1; Table 1). However, the majority of candidates tested in clinical trials so far have been based on different formulations and regions of a handful of antigens identified many years ago, including CSP (51), the liver stage antigen 1 (LSA1) (52), thrombospondin-related antigen (TRAP, also known as sporozoite surface protein 2, SSP2) (53–55), merozoite surface protein 1 (MSP1) (56), MSP2 (57), MSP3 (58), and apical membrane antigen 1 (AMA1) (42, 59). As some of these antigens have shown promising pre-clinical profiles yet limited efficacy in human trials, they have been tested in many different formulations over the years (21, 60). For example, the most advanced blood stage antigens, AMA1 and MSP1, naturally induce protective immune responses (61–63), demonstrated using in vitro inhibition assays (64) and by vaccination of animal models (17, 18, 20, 65). However in humans, only limited clinical efficacy has been observed (66, 67). Similarly, CSP, the major component of the RTS,S vaccine, is the major surface antigen on the sporozoite surface, yet provides only partial and short-lived protection against the blood stage symptoms of malaria following vaccination of human volunteers. It does not protect against infection per se as would be expected by a pre-erythrocytic vaccine and therefore the precise mechanism of protection is not well understood (23, 68). These “historical” vaccine candidates are much further down the development pipeline (21) than more recently identified candidates such as the invasion ligands, the 175 kDa erythrocyte binding antigen (EBA175) (69), reticulocyte binding homolog 5 (RH5) (70, 71), and P. vivax Duffy binding protein (DBP) (72). In recent years, the var2csa variant of PfEMP1, which is the major parasite ligand involved in placental adhesion during pregnancy malaria (73); and the gametocyte antigens, Pfs25 and Pfs48/45 (74) representing promising transmission-blocking targets, have also come to the forefront. The elucidation of the complete parasite genome (75) has further advanced vaccine development by enabling identification of scores of novel antigens. Subsequent antigenic credentialing through functional studies (76), proteomic and immunological screening (63, 77–80), and population genetic analyses (81, 82), has resulted in prioritization of several of these promising candidates for further development.
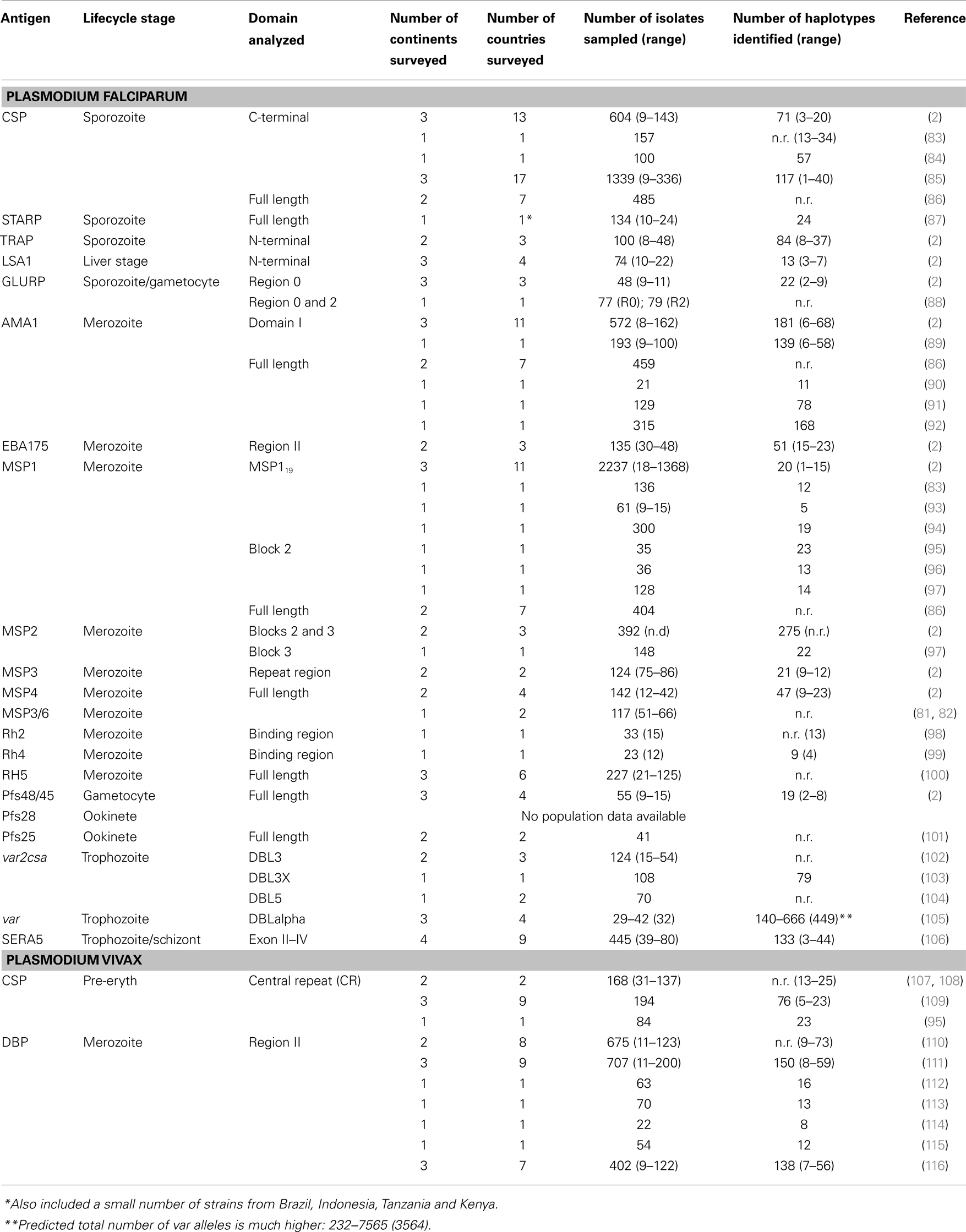
Table 1. Diversity of malaria vaccine candidate antigens currently in clinical trials based on the WHO rainbow tables (50).
Research on potential P. vivax vaccine candidates lags far behind that of P. falciparum. Currently, only two P. vivax vaccine candidates (based on PvCSP and PvDBP) have shown promise in pre-clinical studies and only one candidate has reached Phase 1a clinical trials (50) (Figure 1). If the new goals of the Malaria Vaccine Technology Roadmap are to be realized, significantly more resources need to be invested in identifying and validating promising P. vivax vaccine candidates for progression to clinical trials.
Parasite Diversity: A Major Obstacle to Malaria Vaccine Development
One often quoted explanation for the variable efficacy of subunit malaria vaccine candidates is parasite genetic diversity (2, 9, 12, 37) and the strain-specific nature of immunity to diverse parasite antigens (15, 117, 118). Theoretically, a malaria “strain” can be defined as a parasite variant that is genetically distinct and induces specific immune responses against one or more antigens (Box 1). However, exactly what defines a Plasmodium strain is not fully understood (119) because this definition becomes exceedingly complex when the whole parasite is considered. In the context of subunit vaccines, the term “strain” refers to the parasite isolate from which the vaccine antigen is derived, while the actual genetic variant of that antigen is known as the “allele” or “haplotype” (Box 1). The inclusion of only one allele in a subunit vaccine formulation elicits responses only against similar, cross-reactive alleles (the “serotype,” Box 1) and runs the risk of selection for non-vaccine strains in the vaccinated host population, as discussed in detail below (22). Indeed, natural parasite populations have large numbers of alleles or haplotypes for single copy antigens, such as AMA1 and MSP1 (2, 11) (Table 1). However for PfEMP1, which is encoded by as many as 60 different genes per parasite genome, there are hundreds to thousands of distinct alleles even within local geographic areas <10 km2 (105). Extensive parasite antigenic diversity explains the slow development of naturally acquired immunity (120) with repeated exposure over several years necessary to build up a large repertoire of antibodies to the different serotypes circulating in an endemic area (14, 121). Given the high diversity of the available vaccine candidates (Table 1), a broadly effective malaria vaccine may need to be multivalent, comprising multiple alleles (or haplotypes) for a given polymorphic antigen (12), much like the vaccine approaches used to successfully combat other highly polymorphic pathogens such as influenza A and human papillomavirus.
On the other hand, some Plasmodium antigens are relatively conserved, such as RH5 (100), or have highly conserved functional regions that vaccine-developers may be able to exploit such as the AMA1 receptor-binding pocket (122, 123). Furthermore, antibodies against major surface antigens cross-react with different parasite strains including those from different geographic areas suggesting that conserved epitopes exist (65, 124). It has therefore been proposed that using a panel of peptides containing conserved epitopes would be one approach to induce immune responses that avoid dominant polymorphic epitopes (125).
An important priority in malaria vaccine development is therefore to not only confirm the diversity circulating in the target parasite population but also to understand the contribution of genetic (allelic/haplotypic) diversity to the antigenic (serotype) diversity that is relevant to malaria vaccine design for each candidate antigen (48). Whilst there are indeed multiple diverse alleles of many candidate antigens circulating within distinct populations, not all polymorphisms, will mediate antigenic escape, hence these must be identified and targeted for vaccine design. However, the relationship between allele and serotype has been dissected for only one candidate, AMA1 (126). More rigorous investigation of available candidates as well as the identification of novel relatively conserved antigenic targets is therefore absolutely required to develop a framework for selection and to prioritize antigens for further development as vaccine candidates.
Diversity-Covering Vaccine Approaches
Although well established, the extreme diversity of leading candidate antigens has rarely been considered when developing and testing candidate malaria vaccines [reviewed by Barry et al. (2)]. The majority of subunit vaccine candidates tested in clinical trials have been monovalent. Moreover, all vaccine candidates have been based on alleles from a handful of parasite isolates such as 3D7, FC27, FUP, and FVO for P. falciparum, and Sal1 for P. vivax, that have been propagated for decades in vitro (or in primate models for P. vivax), and poorly reflect the parasite strains circulating in natural populations (2, 110, 127–129). As a result, many malaria vaccine candidates do not adequately cover the diversity observed in natural parasite populations and this could explain the poor clinical efficacy observed in vaccine trials where efficacy endpoints include infection with any strain (21, 66, 127). A multivalent malaria vaccine comprised of multiple serotypes may perform better as it would be designed to protect against a wide range of parasite strains. However, for almost all malaria vaccine candidates, the polymorphisms that define the serotypes and the number of alleles that should be incorporated into a malaria vaccine to cover serotype diversity remain unknown.
Supporting the argument for a multivalent vaccine strategy, vaccine candidates based on a single allele for specific antigens have demonstrated more strongly protective responses when strain-specific endpoints (i.e., infection with a strain carrying the vaccine allele) have been measured as compared to standard endpoints (i.e., infection with any strain). One of the most successful vaccine trials conducted to date was that of the “Combination B” vaccine, conducted in 120 Papua New Guinean children. This vaccine contained only the 3D7 allele of MSP1, MSP2, and the ring-associated erythrocyte antigen (RESA), however, it resulted in a 62% reduction in parasite density in vaccinated children compared to those that received the placebo (22). MSP2 contains a central complex tandem repeat region and many different alleles that vary in size, however all alleles fall into two major families that form different serotypes (3D7 and FC27) (130). Interestingly, at the time of vaccination, the prevalence of the 3D7-type alleles was between 23 and 50% within each of the treatment groups, which could explain the high overall efficacy. Furthermore, vaccinees were less frequently infected and had a lower rate of clinical episodes associated with 3D7-type parasites compared to the control group (22). Similarly, volunteers from Mali vaccinated with FMP2.1, which is based on the 3D7 allele of AMA1, had a much higher risk of non-3D7 infections (64%) (131) than any infection (20%) based on residues in the AMA1 cluster 1 loop (c1L) (66). The results of these trials highlight the danger of vaccine-induced selection pressure and its consequences for morbidity, and strongly argue for developing vaccines covering major serotypes circulating in natural parasite populations (9, 22, 127). The frequency of vaccine or vaccine-serotypes in the target parasite population is also likely to be important for significant vaccine efficacy. The bivalent candidate, AMA1-C1 (containing 3D7 and FVO haplotypes) has demonstrated a lack of protective efficacy against either of the two vaccine alleles in a Phase 2b trial (92). However, this lack of observed efficacy could be explained by a low frequency of these alleles in the target parasite population and the small sample size, with only 44 sequences analyzed for both the vaccine and control groups combined. Even when the analysis of polymorphisms was narrowed to the c1L cluster of polymorphisms, which have been implicated in antigenic escape as the basis of AMA1 serotypes (126), baseline vaccine-allele frequencies were <10% indicating that much larger sample sizes would be required to observe any shift in frequency after vaccination. No other multivalent vaccine trial results are currently available, but several trials are ongoing and the malaria vaccine community awaits the final results with interest.
Multivalent combination vaccines tested in animal models have shown promising and surprising results. Vaccination with combinations of four highly diverse AMA1 alleles was shown to overcome diversity by producing a broader inhibitory response compared to single allele vaccination, thought to have occurred partly by redirecting responses to conserved epitopes (65, 132). This phenomenon, which is analogous to “original antigenic sin,” occurs because abundant, strain-specific AMA1 epitopes vary and are potentially replaced with each new infection, whereas the conserved regions remain constant. Hence, high levels of exposure to conserved epitopes with vaccination or repeated exposure during natural infections may enhance the antibody response against these regions.
Another approach to covering antigenic diversity has been to assemble all available global sequence data for an antigen target and to design a small number of synthetic protein constructs that together cover most of the diversity observed. Phase 1a and 1b trials have begun after promising pre-clinical results for a multivalent vaccine candidate (DiCo), consisting of fusion protein chimeras comprising three synthetic AMA1 molecules covering 97% of the amino acid variability, and these have been shown to elicit stronger antibody responses as a combination than alone (133). This approach has been further evaluated in pre-clinical studies together with a construct containing two allelic variants of the C-terminal 19-kDa region of merozoite surface protein 1 (MSP119) fused to the DiCo construct, and again enhanced antibody responses were induced (134). Clinical trial results are not yet available for the DiCo and MSP119-DiCo combination vaccines but it will be interesting to see whether the diversity-covering approach is more efficacious than the single allele approach.
Predicting Serotypes through Population Genetic Analyses
Population genetic studies are needed to guide vaccine design, by defining the diversity of candidate antigens, to predict polymorphisms that contribute to antigenic diversity (122, 127, 135) and to investigate the geospatial distribution of predicted serotypes (136, 137). Moreover, as a continuous in vitro culture system is yet to be developed for P. vivax, epidemiological studies currently represent an important tool with which to investigate the significance of polymorphism within vaccine candidate antigens (125). This approach has been used to identify correlations between specific polymorphic sites in two leading P. falciparum vaccine candidates, MSP1 and AMA1, with clinical infection (127, 138).
The extensive genetic diversity of malaria vaccine candidate antigens has been demonstrated by many studies investigating genetic polymorphism in samples ranging from small numbers of geographically disparate culture-adapted isolates to large numbers of natural parasite isolates from the same local geographic area or country. These results demonstrate the high numbers of haplotypes found in natural parasite populations for many antigens (Table 1). However, there are fewer haplotypes and vaccine alleles are far more common when individual amino acid polymorphisms or limited “haplotypes” (Box 1) comprising different combinations of amino acid alleles that might form critical epitopes are considered. For example, AMA1, which has 214 amino acid haplotypes in 1 African population, has only 25 serotypes based on the c1L cluster (127), demonstrating that if the haplotype can be refined to represent only antigenic escape polymorphisms, the number of alleles required in a potential multivalent vaccine could be reduced substantially. More recent studies have suggested that the majority of the antigenic escape diversity in AMA1 may even be explained by polymorphism in just one residue (130). Importantly, in vaccine trials, a lack of knowledge of the polymorphisms that mediate antigenic escape would result in an underestimate of strain-specific vaccine efficacy. Population genetic studies are therefore critical to gain more insight into antigenic diversity and to achieve the goal of a broadly efficacious malaria vaccine.
Another important point to remember is that the global P. falciparum and P. vivax populations are structured into geographically distinct subpopulations, therefore, local population-level analyses are required to fully understand diversity that would be relevant to the efficacy of a malaria vaccine in a defined endemic area (139, 140). Given the large number of parasite populations that are likely to exist worldwide it may not be feasible to design vaccines for every target population and therefore a universal approach to cover diversity is needed. Below, we provide a step-by-step guide of how to identify and characterize diversity within candidate antigens that is relevant to malaria vaccine design, and this is further summarized in Figure 2.
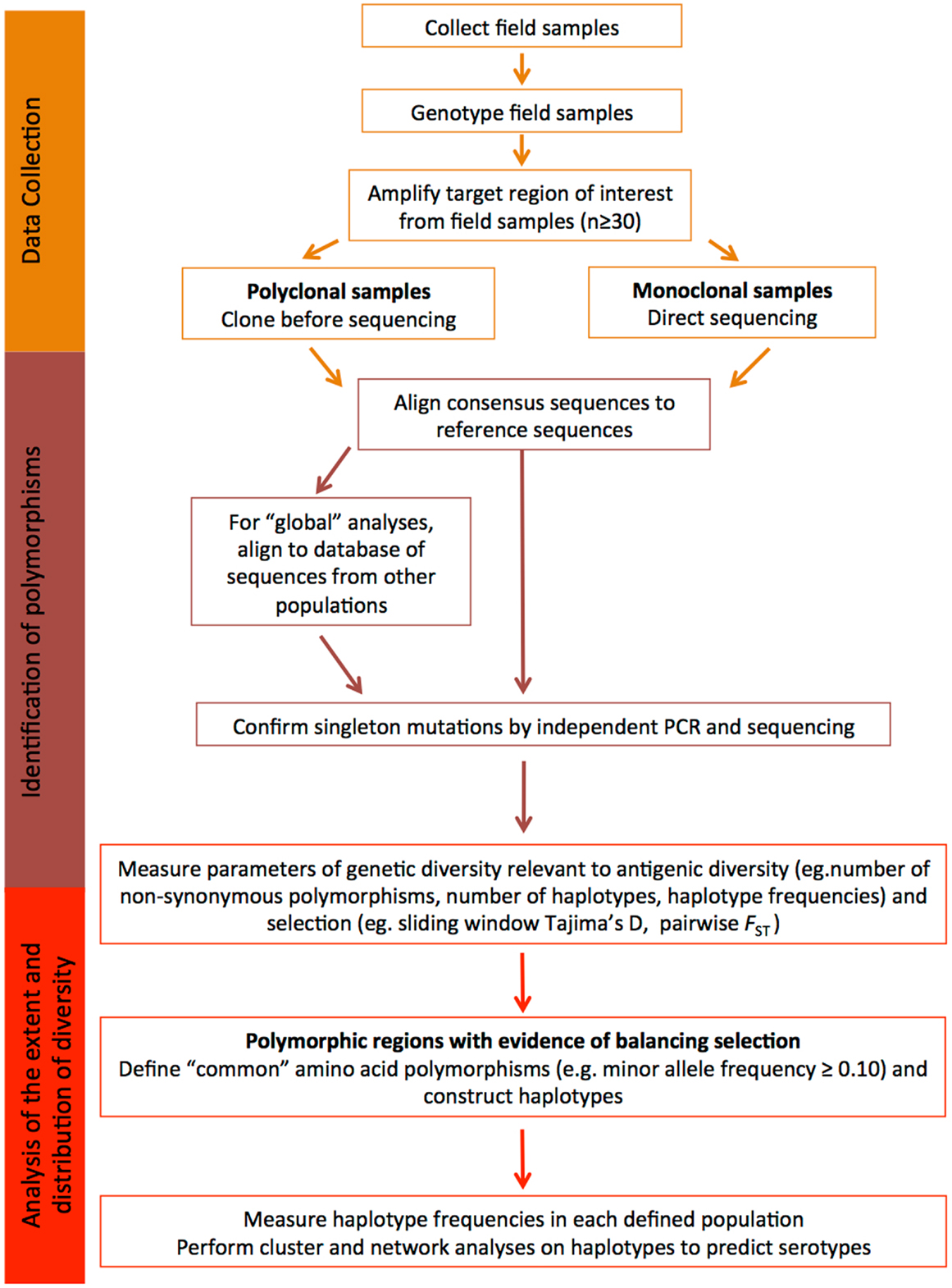
Figure 2. Understanding diversity and predicting serotypes using population genetic analyses. A flow chart describing the step-by-step methodology to define the extent and distribution of parasite diversity and to predict antigenic escape polymorphisms and serotypes (see text for more details).
Data Collection
In order to understand the antigenic diversity impacting on vaccine efficacy and to identify potential serotypes, the target gene or gene region encoding the candidate antigen must be amplified, sequenced, and population genetic analyses completed including the determination of regions under balancing (immune) selection. To accurately estimate natural allele frequencies, it is critical to collect sequence data from samples representing the natural parasite population of a defined geographic area. Use of samples collected in the same geographic area ensures that the diversity of the target sequence is accurately estimated for that region. As the most informative analyses of balancing selection are based on allele frequencies, it is also important that sequence data are obtained from a substantial dataset, at least 30–50 samples (81, 82). Analysis of too few sequences can result in incorrect estimates of diversity, as alleles might be under-represented in very small population samples, and this can result in skewed allele frequencies and diversity estimates.
Prior to PCR amplification, it is essential that parasite isolates are genotyped using highly polymorphic microsatellite markers to determine the number of distinct parasite clones present in the sample (141, 142). Even samples collected from areas of low malaria transmission can contain multiple clones, especially for P. vivax with its frequent relapses (143). If only one clone is present, sequencing can be performed directly from amplicons. If the number of field samples containing a single clone is insufficient, or polyclonal infections are common, PCR products amplified from polyclonal samples can be sub-cloned prior to sequencing. However, sub-cloning often results in artifacts (144), therefore, any novel polymorphisms should be resolved by cloning and sequencing-independent PCR products. Another approach is to perform NextGen sequencing on samples containing multiple clones, once a prohibitively expensive approach that is becoming increasingly affordable. Briefly, amplicons are sheared and adaptors added, generating a library for each sample. Libraries can be sequenced separately or as pools with the addition of sample barcodes. Reads are subsequently mapped to the relevant reference genome. The advantage of this approach is that reads are quantitative and relate to clone abundance in the sample, therefore, haplotypes of the predominant clone(s) can be computationally reconstructed (145).
Identification of Polymorphism
From the raw sequence data, consensus sequences can be obtained and compared to the reference sequence for the defined candidate. Polymorphisms identified in consensus sequences will form the basis of all downstream population genetic analyses. Quality control of sequences is therefore essential, to ensure that polymorphic sites are accurately called and that any ambiguous sites or artifacts are identified. If a clear result cannot be obtained, the sample should be removed from the analysis.
At this point, all high quality consensus sequences from a given population can be compared and basic diversity parameters determined as outlined below. In order to place the dataset in context with the known diversity, published sequences of the target region collected from other natural populations in distinct geographical areas can also be added to the analysis (e.g., from GenBank). Multiple alignments of sequence data are straightforward if sequences are non-repetitive. However, if repeat length polymorphisms, which are common in parasite antigens, or insertions and deletion (indels) are found, then manual realignment needs to be done to ensure that gaps are correctly aligned, which can lead to an overestimate of the number of single nucleotide polymorphisms (SNPs). While repeat regions can be included in population genetic analyses, the expansion and contraction of repeat arrays does not impact on antigenic diversity as dramatically as amino acid changes (22, 131). However, by defining different alleles based on the number of repeats or in the case of indels, the presence or absence of a particular string of nucleotides, it is possible to predict whether such polymorphisms are modulated by immune selection.
Balancing selection, which is a result of immune selection pressure, favors the maintenance of diversity, with alleles at low to medium frequencies within populations, and balanced frequencies between populations (137). Polymorphic sites or regions that show such patterns are predicted to be under the influence of immune selection, and thus contribute to antigenic diversity. For some antigens under strong immune selection, similar alleles or clusters of alleles have been maintained across broad geographic areas (2, 110, 127, 129, 136, 146). The maintenance of moderate frequencies of allele clusters (and even individual alleles) across large geographic distances indicates that they represent distinct serotypes (2, 146).
In contrast, singleton polymorphisms and polymorphic sites with very low minor allele frequencies (MAFs) are indicative of deleterious mutations under purifying selection, or recent polymorphisms yet to increase in frequency. These polymorphisms will only be represented in a very small proportion of the parasite population. As the aim of diversity-covering vaccines is to encompass as many of the alleles (haplotypes) found as possible, several groups have chosen to exclude rare polymorphisms from population genetic analyses of vaccine antigens (110, 129, 145). Another cautionary note is that a large number of singleton polymorphisms in population datasets from the public databases may also indicate PCR artifacts, especially if the methodology includes a cloning step. If the validity of such data cannot be confirmed then it should be discarded from the comparative analyses.
Analysis of the Extent and Distribution of Diversity
In order to determine the diversity of the candidate antigen, both overall and within parasite populations from different geographic areas (e.g., village, district or country), genetic diversity should be estimated for all sequences combined and for each population. Genetic differentiation (e.g., Wrights fixation index, FST) can be measured to determine whether allele frequencies vary and if not, populations from the same region or country can be considered as one population (2). Important diversity parameters to define for each population include the number and type of polymorphisms (SNP, indel, repeat length variation), the number and relative proportions of neutral (synonymous) and amino acid (non-synonymous) polymorphisms, and the number of haplotypes resulting from different combinations of amino acid polymorphisms. All of these statistics can be calculated using freely available population genetic analysis software and an excellent overview of these programs has been published (147). Conversion of polymorphisms to amino acid residues before haplotype definition ensures that complex codons and rare nucleotide polymorphisms within codons containing other more common polymorphisms are included. In any case, it is more appropriate to describe the amino acid haplotypes to predict serotypes, rather than the nucleotide haplotypes. Other more complex statistics such as the nucleotide and haplotype diversity can also be measured as an indication of the number and frequency of polymorphisms and haplotypes in the population, respectively; however, these are of more interest to the population geneticist than the vaccine developer.
To determine whether polymorphic sites are under the influence of balancing (immune) selection, statistical tests such as Tajima’s D can be performed using a sliding window approach, to identify specific domains or clusters of polymorphisms targeted by host immune responses (148). In addition, Wright’s FST, has been applied to investigate whether allele frequencies at specific polymorphic sites, or gene domains, are balanced between populations (137). Polymorphisms under balancing selection and associated with immune escape will be non-synonymous, located in regions with a positive value of Tajima’s D and low inter-population FST values. However, sampling is extremely important as both the Tajima’s D and FST tests are based on allele frequencies. Populations must be of sufficient size so that the sampling error of the estimate is low, and panmictic (randomly mating) from the same geographic region, because any underlying geographic population structure may influence antigen allele frequencies (129, 136, 139, 140). Polymorphisms under balancing selection are also likely to have moderate MAFs, because the maintenance of diversity in the parasite population is advantageous for survival (137). The physical location of all polymorphisms identified as immune targets should then be mapped to the protein structure where possible, to help determine the potential functional relevance and implications of mutation at the site(s) identified, as has been successfully done for AMA1 [e.g., Ref. (126, 127)].
Determining the extent and distribution of immunologically relevant diversity for potential malaria vaccine candidates will help to determine the feasibility of whether a regionally or globally effective vaccine can be developed for the particular target. It is therefore important to focus the analysis to only those polymorphisms with the greatest likelihood of creating antigenic diversity to allow immune escape. Once polymorphisms predicted to be under balancing selection have been identified (see above and Figure 2), the distribution of unique haplotypes representing the predicted serotypes can be analyzed by determining haplotype frequencies in different geographic areas.
Different haplotypes will be related to differing degrees. Using network and clustering analyses, relationships between haplotypes from different populations, clusters of closely related haplotypes, and the extent and distribution of predicted serotypes can be investigated. By identifying the relationships among all of the different haplotypes in this way, the most distantly related alleles can be selected for further analysis or for inclusion in a vaccine to cover diversity. Previous studies have demonstrated the utility of these approaches to identify distinct clusters of alleles as a starting point to predict serotypes (11, 146, 149). These analyses identify the most distinct and common haplotypes and provide a basis upon which to select haplotypes that cover a large proportion of the population-wide diversity for a specific candidate antigen. The outcome of such analyses can help to determine: (i) whether it may be possible to cover all known diversity of the target sequence, (ii) the number of different haplotypes that would need to be included in a vaccine in order to cover diversity and (iii) the predicted efficacy of vaccine candidates. Inclusion of vaccine alleles in the analysis provides a reference point to estimate vaccine allele or serotype frequencies. The results of these analyses provide a simple diversity framework upon which to determine the parameters of allele specific and cross-reactive responses in pre-clinical studies (65, 127, 146) and to measure strain-specific efficacy in clinical trials (131). It is important to note that unlike other approaches that have clearly identified antigenic escape polymorphisms (126, 131), population genetic analyses provide a prediction, are simpler and less expensive, and can reduce the number of polymorphisms (and thus haplotypes) that need to be assessed to confirm their contribution to the serotype.
Defining Antigenic Diversity by Monitoring the Dynamics of Diversity in Natural Parasite Populations
Children living in malaria endemic areas are infected many times and have several episodes of clinical malaria before building up a large repertoire of antibodies that recognize a large number of strains (14, 15). Meanwhile, the rate of turnover of infections increases with age and is thought to be associated with the acquisition of antibodies to an increasing breadth of strains (150). Therefore, strains that children are yet to be exposed to pose a greater risk of clinical illness. Defining the dynamics of clinical infection in the context of parasite antigenic diversity may therefore provide insights into the specific genetic determinants required for antigenic escape, since the risk of a clinical episode due to infection with parasites representing a specific serotype would decrease after being exposed to that serotype (Figure 3A) (127, 138). Furthermore, analyses of strain-specific antibodies in conjunction with such analyses would confirm the status of immunity at a particular time point and allow associations to be made between gaps in the antibody repertoire and risk of a clinical episode (62, 98, 99, 151). Longitudinal studies in endemic regions that monitor children at regular time intervals for the presence of specific antibodies or the turnover of alleles at antigen loci could therefore be harnessed to identify the genetic determinants of antigenic diversity.
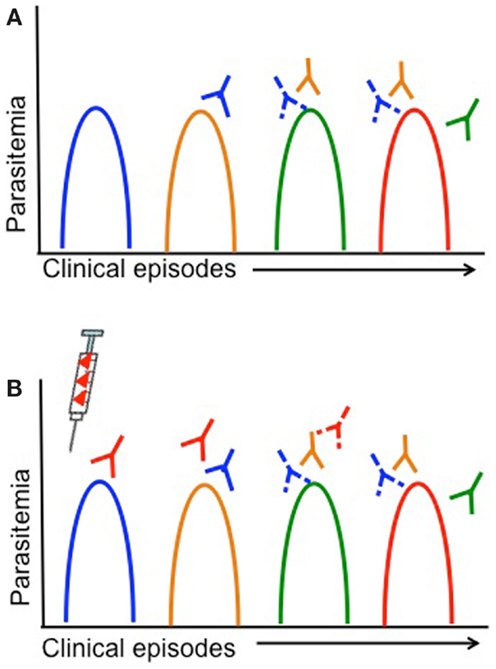
Figure 3. Antigenic diversity, clinical malaria, and vaccine efficacy are shown. Simplified overview of the impact of parasite antigenic diversity on the dynamics of natural infection and the efficacy of vaccines. Peaks in parasitemia correspond with different clinical episodes and colors indicate different serotypes. Strain-specific acquired antibodies are shown in corresponding colors some time after each clinical episode. Solid lines represent a strong antibody response, while dashed lines represent limited antibody responses. (A) Dynamics of natural infection with recurrent episodes of clinical malaria in an individual that acquires only strain-specific antibodies after being infected. As individuals experience different strains through natural infection (or vaccination), they acquire strain-specific antibodies and have a lower risk of having a clinical episode due to the same strain. (B) Lack of vaccine efficacy in a vaccinated individual due to antigenic diversity. The syringe indicates vaccination with a single serotype (red). If a single-strain vaccine is given at baseline, individuals are more likely to experience clinical episodes due to other strains (blue, yellow, green) than the vaccine strain, until antibody responses decrease.
Measuring Strain-Specific Efficacy in Vaccine Trials
Vaccines based on a single allele of the candidate antigen elicit strain-specific antibodies, so that vaccinated individuals continue to be at risk of infection with different strains but risk of infection with the vaccine strain is lower (Figure 3B) (22, 66, 131). Vaccine trials with strain-specific endpoints (i.e., infection with a particular parasite genotype) therefore represent a major opportunity to characterize the genetic determinants responsible for immune escape; however, this has been rarely attempted. Given the high diversity of most malaria vaccine candidates and the limited efficacy of single-strain vaccine candidates, for any candidate that elicits strain-specific responses it will be essential to measure strain-specific endpoints to ensure that antigenic escape is explored in vaccine testing. The advent of high throughput genotyping approaches (142, 152) has reduced the effort and funding required for molecular epidemiological studies, and therefore as long as relevant expertise is available, these investigations should be relatively simple to incorporate into a vaccine trial.
The primary efficacy endpoint in clinical trials of malaria vaccines has included a range of measurements such as time to first infection, occurrence of a clinical episode (mild and severe), and parasite density. However, only a few malaria vaccine trials have determined whether efficacy against vaccine or closely related strains has been achieved (22, 92, 131). Below, we cover some molecular epidemiological analyses that could be considered in the different stages of a vaccine trial.
Vaccine Trial Reconnaissance
The low prevalence of vaccine alleles within natural populations has been proposed to limit vaccine efficacy (2, 11, 129, 138). Furthermore, a low prevalence of the vaccine allele will make it extremely difficult to measure allele specific efficacy, since changes may be very small and therefore require exceedingly large sample sizes (92). Therefore, even before a vaccine trial begins in a particular geographic area, molecular epidemiological surveys will be important to assess baseline population genetic characteristics, and in particular the allele- (or if known, the serotype-) frequencies to ensure that the vaccine to be tested is representative of the target parasite population.
Strain-Specific Endpoints
In addition to measuring strain-specific endpoints by determining the time to first clinical episode with vaccine and non-vaccine alleles, the analysis of outcomes in control and vaccine groups provide an opportunity for more detailed analyses that can provide key insights into the complexities of parasite antigenic diversity (92, 127, 153). In principle, the same analyses can be done in longitudinal studies to achieve the same goal. These analyses will provide further functional evidence of antigenic escape polymorphisms, thus allowing refinement of the serotype. Different types of analyses that have been used to measure strain-specific efficacy in vaccine trials include:
(i) measuring strain-specific protection by assessing risk of clinical episode associated with parasites carrying the vaccine-allele compared to those carrying any non-vaccine allele;
(ii) measuring cross-strain protection by assessing risk of clinical infection with parasites carrying the vaccine-allele compared to those carrying different non-vaccine alleles;
(iii) measuring vaccine-mediated selection by:
a. comparison of allele or haplotype frequencies before and after vaccination in the different vaccine groups;
b. comparison of the incidence of infection with parasites carrying different haplotypes and individual polymorphisms before and after vaccination;
(iv) assessment of vaccine-mediated strain-specific natural immunity by measuring antibodies to vaccine alleles in vaccine and control groups.
Vaccine-Mediated Selection and Antigenic Escape Mutants
The development and licensing of an effective malaria vaccine will require sustained and intensive surveillance to monitor for vaccine-mediated selection of non-vaccine alleles and to ensure that vaccine alleles are common enough for vaccines to remain effective. Regular population genetic surveys will be required in order to monitor allele frequencies, to ensure that alleles contained within the vaccine continue to represent the diversity of the parasite population and that new, potential antigenic escape mutants have not emerged. A genetic surveillance system would be difficult and costly to implement, but the development of low cost rapid assays to quickly genotype parasite isolates would facilitate such an approach (152). The need for simple and informative surveillance tools also highlights the importance of gaining knowledge into the precise determinants of antigenic diversity in any antigen to be included in a licensed malaria vaccine.
Conclusion
The trials and tribulations of malaria vaccine development have reached a critical juncture. The first licensed malaria vaccine is almost at hand and children in African countries stand to benefit greatly from its availability. However, this vaccine is only partially effective against the symptoms of malaria, not infection, and provides only short-lived protection (23). As a result of the success of the reinvigorated global malaria eradication program, many previously highly endemic countries no longer have high burdens of disease. By 2030, the goal is to have a second-generation vaccine(s) that can provide broad and long-lived protection against multiple species and diverse strains. Parasite antigenic diversity, one of the major reasons for the failures of past candidate malaria vaccines, remains a barrier to the efficacy of subunit and potentially even whole parasite vaccine approaches (48), but there is light at the end of the tunnel: this review demonstrates how it may be possible to overcome parasite antigenic diversity. Successful identification of the critical genetic determinants of the serotype for one leading vaccine candidate, AMA1 (126, 127, 131) and proof that multivalent vaccines representing the majority of the diversity of this antigen can generate broad antibody responses further support the development of effective multi-allele vaccines and suggest that antigenic diversity in malaria may be overcome (65, 132). By consolidating the vast knowledge gained on the genetic diversity of candidate antigens, by harnessing high throughput genotyping tools and with careful design of longitudinal studies and vaccine trials, it is highly likely that it will also be possible to identify the critical genetic determinants underlying antigenic diversity (i.e., the serotype) for any candidate malaria vaccine antigen.
Author Contributions
Alyssa E. Barry and Alicia Arnott wrote the paper, collated data, and prepared figures.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to all of our colleagues and mentors who have contributed to discussions about the concepts outlined in this review over the last several years, in particular, J. Reeder, J. Beeson, M. Good, and I. Mueller. The authors’ research is supported by Project Grants awarded to Alyssa E. Barry by the National Health and Medical Research Council of Australia (No. 488211, 1005653 and 1003825). The work was made possible through Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS.
References
2. Barry AE, Schultz L, Buckee CO, Reeder JC. Contrasting population structures of the genes encoding ten leading vaccine-candidate antigens of the human malaria parasite, Plasmodium falciparum. PLoS One (2009) 4(12):e8497. doi: 10.1371/journal.pone.0008497
3. Roll Back Malaria Partnership. The Global Malaria Action Plan for a Malaria Free World. Geneva: Roll Back Malaria Partnership (2008).
4. Gething PW, Elyazar IR, Moyes CL, Smith DL, Battle KE, Guerra CA, et al. A long neglected world malaria map: Plasmodium vivax endemicity in 2010. PLoS Negl Trop Dis (2012) 6(9):e1814. doi:10.1371/journal.pntd.0001814
5. Cotter C, Sturrock HJ, Hsiang MS, Liu J, Phillips AA, Hwang J, et al. The changing epidemiology of malaria elimination: new strategies for new challenges. Lancet (2013) 382(9895):900–11. doi:10.1016/S0140-6736(13)60310-4
6. Mueller I, Galinski MR, Baird JK, Carlton JM, Kochar DK, Alonso PL, et al. Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect Dis (2009) 9(9):555–66. doi:10.1016/S1473-3099(09)70177-X
8. Alonso PL, Brown G, Arevalo-Herrera M, Binka F, Chitnis C, Collins F, et al. A research agenda to underpin malaria eradication. PLoS Med (2011) 8(1):e1000406. doi:10.1371/journal.pmed.1000406
9. Takala SL, Plowe CV. Genetic diversity and malaria vaccine design, testing and efficacy: preventing and overcoming “vaccine resistant malaria”. Parasite Immunol (2009) 31(9):560–73. doi:10.1111/j.1365-3024.2009.01138.x
10. Riley EM, Stewart VA. Immune mechanisms in malaria: new insights in vaccine development. Nat Med (2013) 19(2):168–78. doi:10.1038/nm.3083
11. Barry AE, Beeson J, Reeder JC, Fowkes F, Arnott A. Using population genetics to guide malaria vaccine design. In: Okwa APO, editor. Malaria Parasites. Rijeka: InTech (2012). p. 231–64.
12. Richards JS, Beeson JG. The future for blood-stage vaccines against malaria. Immunol Cell Biol (2009) 87(5):377–90. doi:10.1038/icb.2009.27
13. Koch R. Dritter Bericht uber die Thatigkeit der Malaria-Expedition. Dtsch Med Wochenschr (1900) 26(17):281–4. doi:10.1055/s-0029-1203823
14. Marsh K, Kinyanjui S. Immune effector mechanisms in malaria. Parasite Immunol (2006) 28(1–2):51–60. doi:10.1111/j.1365-3024.2006.00808.x
15. Doolan DL, Dobano C, Baird JK. Acquired immunity to malaria. Clin Microbiol Rev (2009) 22(1):13–36. doi:10.1128/CMR.00025-08
16. Cohen S, Mc GI, Carrington S. Gamma-globulin and acquired immunity to human malaria. Nature (1961) 192:733–7. doi:10.1038/192733a0
17. Darko CA, Angov E, Collins WE, Bergmann-Leitner ES, Girouard AS, Hitt SL, et al. The clinical-grade 42-kilodalton fragment of merozoite surface protein 1 of Plasmodium falciparum strain FVO expressed in Escherichia coli protects Aotus nancymai against challenge with homologous erythrocytic-stage parasites. Infect Immun (2005) 73(1):287–97. doi:10.1128/IAI.73.1.287-297.2005
18. Dutta S, Sullivan JS, Grady KK, Haynes JD, Komisar J, Batchelor AH, et al. High antibody titer against apical membrane antigen-1 is required to protect against malaria in the Aotus model. PLoS One (2009) 4(12):e8138. doi:10.1371/journal.pone.0008138
19. Gentil F, Bargieri DY, Leite JA, Francoso KS, Patricio MB, Espindola NM, et al. A recombinant vaccine based on domain II of Plasmodium vivax apical membrane antigen 1 induces high antibody titres in mice. Vaccine (2010) 28(38):6183–90. doi:10.1016/j.vaccine.2010.07.017
20. Stowers AW, Kennedy MC, Keegan BP, Saul A, Long CA, Miller LH. Vaccination of monkeys with recombinant Plasmodium falciparum apical membrane antigen 1 confers protection against blood-stage malaria. Infect Immun (2002) 70(12):6961–7. doi:10.1128/IAI.70.12.6961-6967.2002
21. Schwartz L, Brown GV, Genton B, Moorthy VS. A review of malaria vaccine clinical projects based on the WHO rainbow table. Malar J (2012) 11:11. doi:10.1186/1475-2875-11-11
22. Genton B, Betuela I, Felger I, Al-Yaman F, Anders RF, Saul A, et al. A recombinant blood-stage malaria vaccine reduces Plasmodium falciparum density and exerts selective pressure on parasite populations in a phase 1-2b trial in Papua New Guinea. J Infect Dis (2002) 185(6):820–7. doi:10.1086/339342
23. Agnandji ST, Lell B, Soulanoudjingar SS, Fernandes JF, Abossolo BP, Conzelmann C, et al. First results of phase 3 trial of RTS,S/AS01 malaria vaccine in African children. N Engl J Med (2011) 365(20):1863–75. doi:10.1056/NEJMoa1102287
24. Sirima SB, Cousens S, Druilhe P. Protection against malaria by MSP3 candidate vaccine. N Engl J Med (2011) 365(11):1062–4. doi:10.1056/NEJMc1100670
25. Laurens MB, Thera MA, Coulibaly D, Ouattara A, Kone AK, Guindo AB, et al. Extended safety, immunogenicity and efficacy of a blood-stage malaria vaccine in malian children: 24-month follow-up of a randomized, double-blinded phase 2 trial. PLoS One (2013) 8(11):e79323. doi:10.1371/journal.pone.0079323
26. Clyde DF, Most H, McCarthy VC, Vanderberg JP. Immunization of man against sporozite-induced falciparum malaria. Am J Med Sci (1973) 266(3):169–77. doi:10.1097/00000441-197312000-00001
27. Hoffman SL, Goh LM, Luke TC, Schneider I, Le TP, Doolan DL, et al. Protection of humans against malaria by immunization with radiation-attenuated Plasmodium falciparum sporozoites. J Infect Dis (2002) 185(8):1155–64. doi:10.1086/339409
28. Nussenzweig V, Nussenzweig RS. Circumsporozoite proteins of malaria parasites. Cell (1985) 42(2):401–3. doi:10.1016/0092-8674(85)90093-5
29. Malaria Vaccine Funders Group. Malaria Vaccine Technology Roadmap. Geneva: World Health Organization (2006).
30. Otieno L. Efficacy of RTS,S/AS01 vaccine candidate against malaria in African infants and children 8 months post-primary vaccination series: a phase III randomized, double-blind controlled trial. Multilateral Initiative on Malaria Pan Africa Conference. Durban (2013).
31. Malaria Vaccine Funders Group. Malaria Vaccine Technology Roadmap. Geneva: World Health Organization (2013).
32. Doolan DL, Aguiar JC, Weiss WR, Sette A, Felgner PL, Regis DP, et al. Utilization of genomic sequence information to develop malaria vaccines. J Exp Biol (2003) 206(Pt 21):3789–802. doi:10.1242/jeb.00615
33. Roestenberg M, McCall M, Hopman J, Wiersma J, Luty AJ, van Gemert GJ, et al. Protection against a malaria challenge by sporozoite inoculation. N Engl J Med (2009) 361(5):468–77. doi:10.1056/NEJMoa0805832
34. Teirlinck AC, McCall MB, Roestenberg M, Scholzen A, Woestenenk R, de Mast Q, et al. Longevity and composition of cellular immune responses following experimental Plasmodium falciparum malaria infection in humans. PLoS Pathog (2011) 7(12):e1002389. doi:10.1371/journal.ppat.1002389
35. Butler NS, Vaughan AM, Harty JT, Kappe SH. Whole parasite vaccination approaches for prevention of malaria infection. Trends Immunol (2012) 33(5):247–54. doi:10.1016/j.it.2012.02.001
36. Hill AV. Vaccines against malaria. Philos Trans R Soc Lond B Biol Sci (2011) 366(1579):2806–14. doi:10.1098/rstb.2011.0091
37. Stanisic DI, Barry AE, Good MF. Escaping the immune system: how the malaria parasite makes vaccine development a challenge. Trends Parasitol (2013) 29(12):612–22. doi:10.1016/j.pt.2013.10.001
38. Singh S, Miura K, Zhou H, Muratova O, Keegan B, Miles A, et al. Immunity to recombinant Plasmodium falciparum merozoite surface protein 1 (MSP1): protection in Aotus nancymai monkeys strongly correlates with anti-MSP1 antibody titer and in vitro parasite-inhibitory activity. Infect Immun (2006) 74(8):4573–80. doi:10.1128/IAI.01679-05
39. Dent AE, Bergmann-Leitner ES, Wilson DW, Tisch DJ, Kimmel R, Vulule J, et al. Antibody-mediated growth inhibition of Plasmodium falciparum: relationship to age and protection from parasitemia in Kenyan children and adults. PLoS One (2008) 3(10):e3557. doi:10.1371/journal.pone.0003557
40. King CL, Michon P, Shakri AR, Marcotty A, Stanisic D, Zimmerman PA, et al. Naturally acquired Duffy-binding protein-specific binding inhibitory antibodies confer protection from blood-stage Plasmodium vivax infection. Proc Natl Acad Sci U S A (2008) 105(24):8363–8. doi:10.1073/pnas.0800371105
41. McCallum FJ, Persson KE, Mugyenyi CK, Fowkes FJ, Simpson JA, Richards JS, et al. Acquisition of growth-inhibitory antibodies against blood-stage Plasmodium falciparum. PLoS One (2008) 3(10):e3571. doi:10.1371/journal.pone.0003571
42. Deans JA, Knight AM, Jean WC, Waters AP, Cohen S, Mitchell GH. Vaccination trials in rhesus monkeys with a minor, invariant, Plasmodium knowlesi 66 kD merozoite antigen. Parasite Immunol (1988) 10(5):535–52. doi:10.1111/j.1365-3024.1988.tb00241.x
43. Renia L, Ling IT, Marussig M, Miltgen F, Holder AA, Mazier D. Immunization with a recombinant C-terminal fragment of Plasmodium yoelii merozoite surface protein 1 protects mice against homologous but not heterologous P. yoelii sporozoite challenge. Infect Immun (1997) 65(11):4419–23.
44. Biswas S, Spencer AJ, Forbes EK, Gilbert SC, Holder AA, Hill AV, et al. Recombinant viral-vectored vaccines expressing Plasmodium chabaudi AS apical membrane antigen 1: mechanisms of vaccine-induced blood-stage protection. J Immunol (2012) 188(10):5041–53. doi:10.4049/jimmunol.1101106
45. Lyon JA, Angov E, Fay MP, Sullivan JS, Girourd AS, Robinson SJ, et al. Protection induced by Plasmodium falciparum MSP1(42) is strain-specific, antigen and adjuvant dependent, and correlates with antibody responses. PLoS One (2008) 3(7):e2830. doi:10.1371/journal.pone.0002830
46. Birkett AJ, Moorthy VS, Loucq C, Chitnis CE, Kaslow DC. Malaria vaccine R&D in the decade of vaccines: breakthroughs, challenges and opportunities. Vaccine (2013) 31(Suppl 2):B233–43. doi:10.1016/j.vaccine.2013.02.040
47. Sultan AA, Thathy V, Nussenzweig V, Menard R. Green fluorescent protein as a marker in Plasmodium berghei transformation. Infect Immun (1999) 67(5):2602–6.
48. The malERA Consultative Group on Vaccines. A research agenda for malaria eradication: vaccines. PLoS Med (2011) 8(1):e1000398. doi:10.1371/journal.pmed.1000398
49. Hviid L. The role of Plasmodium falciparum variant surface antigens in protective immunity and vaccine development. Hum Vaccin (2010) 6(1):84–9. doi:10.4161/hv.6.1.9602
50. World Health Organisation. Malaria Vaccine Rainbow Tables (2013). Available from: http://www.who.int/vaccine_research/links/Rainbow/en/index.html
51. Yoshida N, Nussenzweig RS, Potocnjak P, Nussenzweig V, Aikawa M. Hybridoma produces protective antibodies directed against the sporozoite stage of malaria parasite. Science (1980) 207(4426):71–3. doi:10.1126/science.6985745
52. Guerin-Marchand C, Druilhe P, Galey B, Londono A, Patarapotikul J, Beaudoin RL, et al. A liver-stage-specific antigen of Plasmodium falciparum characterized by gene cloning. Nature (1987) 329(6135):164–7. doi:10.1038/329164a0
53. Robson KJ, Hall JR, Jennings MW, Harris TJ, Marsh K, Newbold CI, et al. A highly conserved amino-acid sequence in thrombospondin, properdin and in proteins from sporozoites and blood stages of a human malaria parasite. Nature (1988) 335(6185):79–82. doi:10.1038/335079a0
54. Rogers WO, Malik A, Mellouk S, Nakamura K, Rogers MD, Szarfman A, et al. Characterization of Plasmodium falciparum sporozoite surface protein 2. Proc Natl Acad Sci U S A (1992) 89(19):9176–80. doi:10.1073/pnas.89.19.9176
55. Cowan G, Krishna S, Crisanti A, Robson K. Expression of thrombospondin-related anonymous protein in Plasmodium falciparum sporozoites. Lancet (1992) 339(8806):1412–3. doi:10.1016/0140-6736(92)91229-2
56. Holder AA, Blackman MJ, Burghaus PA, Chappel JA, Ling IT, McCallum-Deighton N, et al. A malaria merozoite surface protein (MSP1)-structure, processing and function. Mem Inst Oswaldo Cruz (1992) 87(Suppl 3):37–42. doi:10.1590/S0074-02761992000700004
57. Smythe JA, Coppel RL, Brown GV, Ramasamy R, Kemp DJ, Anders RF. Identification of two integral membrane proteins of Plasmodium falciparum. Proc Natl Acad Sci U S A (1988) 85(14):5195–9. doi:10.1073/pnas.85.14.5195
58. Oeuvray C, Bouharoun-Tayoun H, Gras-Masse H, Bottius E, Kaidoh T, Aikawa M, et al. Merozoite surface protein-3: a malaria protein inducing antibodies that promote Plasmodium falciparum killing by cooperation with blood monocytes. Blood (1994) 84(5):1594–602.
59. Hodder AN, Crewther PE, Matthew ML, Reid GE, Moritz RL, Simpson RJ, et al. The disulfide bond structure of Plasmodium apical membrane antigen-1. J Biol Chem (1996) 271(46):29446–52. doi:10.1074/jbc.271.46.29446
60. Genton B, Reed ZH. Asexual blood-stage malaria vaccine development: facing the challenges. Curr Opin Infect Dis (2007) 20(5):467–75. doi:10.1097/QCO.0b013e3282dd7a29
61. Fowkes FJ, Richards JS, Simpson JA, Beeson JG. The relationship between anti-merozoite antibodies and incidence of Plasmodium falciparum malaria: a systematic review and meta-analysis. PLoS Med (2010) 7(1):e1000218. doi:10.1371/journal.pmed.1000218
62. Stanisic DI, Richards JS, McCallum FJ, Michon P, King CL, Schoepflin S, et al. Immunoglobulin G subclass-specific responses against Plasmodium falciparum merozoite antigens are associated with control of parasitemia and protection from symptomatic illness. Infect Immun (2009) 77(3):1165–74. doi:10.1128/IAI.01129-08
63. Richards JS, Arumugam TU, Reiling L, Healer J, Hodder AN, Fowkes FJ, et al. Identification and prioritization of merozoite antigens as targets of protective human immunity to Plasmodium falciparum malaria for vaccine and biomarker development. J Immunol (2013) 191(2):795–809. doi:10.4049/jimmunol.1300778
64. Baum J, Chen L, Healer J, Lopaticki S, Boyle M, Triglia T, et al. Reticulocyte-binding protein homologue 5 – an essential adhesin involved in invasion of human erythrocytes by Plasmodium falciparum. Int J Parasitol (2009) 39(3):371–80. doi:10.1016/j.ijpara.2008.10.006
65. Dutta S, Dlugosz LS, Drew DR, Ge X, Ababacar D, Rovira YI, et al. Overcoming antigenic diversity by enhancing the immunogenicity of conserved epitopes on the malaria vaccine candidate apical membrane antigen-1. PLoS Pathog (2013) 9(12):e1003840. doi:10.1371/journal.ppat.1003840
66. Thera MA, Doumbo OK, Coulibaly D, Laurens MB, Ouattara A, Kone AK, et al. A field trial to assess a blood-stage malaria vaccine. N Engl J Med (2011) 365(11):1004–13. doi:10.1056/NEJMoa1008115
67. Ogutu BR, Apollo OJ, McKinney D, Okoth W, Siangla J, Dubovsky F, et al. Blood stage malaria vaccine eliciting high antigen-specific antibody concentrations confers no protection to young children in Western Kenya. PLoS One (2009) 4(3):e4708. doi:10.1371/journal.pone.0004708
68. Fowkes FJ, Simpson JA, Beeson JG. Implications of the licensure of a partially efficacious malaria vaccine on evaluating second-generation vaccines. BMC Med (2013) 11:232. doi:10.1186/1741-7015-11-232
69. El Sahly HM, Patel SM, Atmar RL, Lanford TA, Dube T, Thompson D, et al. Safety and immunogenicity of a recombinant nonglycosylated erythrocyte binding antigen 175 region II malaria vaccine in healthy adults living in an area where malaria is not endemic. Clin Vaccine Immunol (2010) 17(10):1552–9. doi:10.1128/CVI.00082-10
70. Douglas AD, Williams AR, Illingworth JJ, Kamuyu G, Biswas S, Goodman AL, et al. The blood-stage malaria antigen PfRH5 is susceptible to vaccine-inducible cross-strain neutralizing antibody. Nat Commun (2011) 2:601. doi:10.1038/ncomms1615
71. Crosnier C, Bustamante LY, Bartholdson SJ, Bei AK, Theron M, Uchikawa M, et al. Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum. Nature (2011) 480(7378):534–7. doi:10.1038/nature10606
72. Ntumngia FB, Schloegel J, McHenry AM, Barnes SJ, George MT, Kennedy S, et al. Immunogenicity of single versus mixed allele vaccines of Plasmodium vivax Duffy binding protein region II. Vaccine (2013) 31(40):4382–8. doi:10.1016/j.vaccine.2013.07.002
73. Salanti A, Staalsoe T, Lavstsen T, Jensen AT, Sowa MP, Arnot DE, et al. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol Microbiol (2003) 49(1):179–91. doi:10.1046/j.1365-2958.2003.03570.x
74. Rener J, Graves PM, Carter R, Williams JL, Burkot TR. Target antigens of transmission-blocking immunity on gametes of Plasmodium falciparum. J Exp Med (1983) 158(3):976–81. doi:10.1084/jem.158.3.976
75. Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature (2002) 419(6906):498–511. doi:10.1038/nature01097
76. Taechalertpaisarn T, Crosnier C, Bartholdson SJ, Hodder AN, Thompson J, Bustamante LY, et al. Biochemical and functional analysis of two Plasmodium falciparum blood-stage 6-cys proteins: P12 and P41. PLoS One (2012) 7(7):e41937. doi:10.1371/journal.pone.0041937
77. Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, Haynes JD, et al. A proteomic view of the Plasmodium falciparum life cycle. Nature (2002) 419(6906):520–6. doi:10.1038/nature01107
78. Sanders PR, Gilson PR, Cantin GT, Greenbaum DC, Nebl T, Carucci DJ, et al. Distinct protein classes including novel merozoite surface antigens in Raft-like membranes of Plasmodium falciparum. J Biol Chem (2005) 280(48):40169–76. doi:10.1074/jbc.M509631200
79. Doolan DL, Mu Y, Unal B, Sundaresh S, Hirst S, Valdez C, et al. Profiling humoral immune responses to P. falciparum infection with protein microarrays. Proteomics (2008) 8(22):4680–94. doi:10.1002/pmic.200800194
80. Persson KE, Fowkes FJ, McCallum FJ, Gicheru N, Reiling L, Richards JS, et al. Erythrocyte-binding antigens of Plasmodium falciparum are targets of human inhibitory antibodies and function to evade naturally acquired immunity. J Immunol (2013) 191(2):785–94. doi:10.4049/jimmunol.1300444
81. Tetteh KK, Stewart LB, Ochola LI, Amambua-Ngwa A, Thomas AW, Marsh K, et al. Prospective identification of malaria parasite genes under balancing selection. PLoS One (2009) 4(5):e5568. doi:10.1371/journal.pone.0005568
82. Ochola LI, Tetteh KK, Stewart LB, Riitho V, Marsh K, Conway DJ. Allele frequency-based and polymorphism-versus-divergence indices of balancing selection in a new filtered set of polymorphic genes in Plasmodium falciparum. Mol Biol Evol (2010) 27(10):2344–51. doi:10.1093/molbev/msq119
83. Kariuki SK, Njunge J, Muia A, Muluvi G, Gatei W, Ter Kuile F, et al. Effect of malaria transmission reduction by insecticide-treated bed nets (ITNs) on the genetic diversity of Plasmodium falciparum merozoite surface protein (MSP-1) and circumsporozoite (CSP) in Western Kenya. Malar J (2013) 12:295. doi:10.1186/1475-2875-12-295
84. Bailey JA, Mvalo T, Aragam N, Weiser M, Congdon S, Kamwendo D, et al. Use of massively parallel pyrosequencing to evaluate the diversity of and selection on Plasmodium falciparum csp T-cell epitopes in Lilongwe, Malawi. J Infect Dis (2012) 206(4):580–7. doi:10.1093/infdis/jis329
85. Zeeshan M, Alam MT, Vinayak S, Bora H, Tyagi RK, Alam MS, et al. Genetic variation in the Plasmodium falciparum circumsporozoite protein in India and its relevance to RTS,S malaria vaccine. PLoS One (2012) 7(8):e43430. doi:10.1371/journal.pone.0043430
86. Tanabe K, Mita T, Palacpac NM, Arisue N, Tougan T, Kawai S, et al. Within-population genetic diversity of Plasmodium falciparum vaccine candidate antigens reveals geographic distance from a Central sub-Saharan African origin. Vaccine (2013) 31(9):1334–9. doi:10.1016/j.vaccine.2012.12.039
87. Jongwutiwes S, Putaporntip C, Karnchaisri K, Seethamchai S, Hongsrimuang T, Kanbara H. Positive selection on the Plasmodium falciparum sporozoite threonine-asparagine-rich protein: analysis of isolates mainly from low endemic areas. Gene (2008) 410(1):139–46. doi:10.1016/j.gene.2007.12.006
88. Pratt-Riccio LR, Perce-da-Silva Dde S, Lima-Junior Jda C, Theisen M, Santos F, Daniel-Ribeiro CT, et al. Genetic polymorphisms in the glutamate-rich protein of Plasmodium falciparum field isolates from a malaria-endemic area of Brazil. Mem Inst Oswaldo Cruz (2013) 108(4):523–8. doi:10.1590/S0074-02762013000400022
89. Basu M, Maji AK, Mitra M, Sengupta S. Natural selection and population genetic structure of domain-I of Plasmodium falciparum apical membrane antigen-1 in India. Infect Genet Evol (2013) 18:247–56. doi:10.1016/j.meegid.2013.05.015
90. Mehrizi AA, Sepehri M, Karimi F, Djadid ND, Zakeri S. Population genetics, sequence diversity and selection in the gene encoding the Plasmodium falciparum apical membrane antigen 1 in clinical isolates from the south-east of Iran. Infect Genet Evol (2013) 17:51–61. doi:10.1016/j.meegid.2013.03.042
91. Osier FH, Weedall GD, Verra F, Murungi L, Tetteh KK, Bull P, et al. Allelic diversity and naturally acquired allele-specific antibody responses to Plasmodium falciparum apical membrane antigen 1 in Kenya. Infect Immun (2010) 78(11):4625–33. doi:10.1128/IAI.00576-10
92. Ouattara A, Mu J, Takala-Harrison S, Saye R, Sagara I, Dicko A, et al. Lack of allele-specific efficacy of a bivalent AMA1 malaria vaccine. Malar J (2010) 9:175. doi:10.1186/1475-2875-9-175
93. Simpalipan P, Pattaradilokrat S, Siripoon N, Seugorn A, Kaewthamasorn M, Butcher RD, et al. Diversity and population structure of Plasmodium falciparum in Thailand based on the spatial and temporal haplotype patterns of the C-terminal 19-kDa domain of merozoite surface protein-1. Malar J (2014) 13(1):54. doi:10.1186/1475-2875-13-54
94. Pan D, Hu J, Ma Q, Pan W, Li M. Diversity and prevalence of the C-terminal region of Plasmodium falciparum merozoite surface protein 1 in China. Acta Trop (2010) 116(3):200–5. doi:10.1016/j.actatropica.2010.08.004
95. Lopez AC, Ortiz A, Coello J, Sosa-Ochoa W, Torres RE, Banegas EI, et al. Genetic diversity of Plasmodium vivax and Plasmodium falciparum in Honduras. Malar J (2012) 11:391. doi:10.1186/1475-2875-11-391
96. Yuan L, Zhao H, Wu L, Li X, Parker D, Xu S, et al. Plasmodium falciparum populations from northeastern Myanmar display high levels of genetic diversity at multiple antigenic loci. Acta Trop (2013) 125(1):53–9. doi:10.1016/j.actatropica.2012.09.008
97. Kang JM, Moon SU, Kim JY, Cho SH, Lin K, Sohn WM, et al. Genetic polymorphism of merozoite surface protein-1 and merozoite surface protein-2 in Plasmodium falciparum field isolates from Myanmar. Malar J (2010) 9:131. doi:10.1186/1475-2875-9-131
98. Reiling L, Richards JS, Fowkes FJ, Barry AE, Triglia T, Chokejindachai W, et al. Evidence that the erythrocyte invasion ligand PfRh2 is a target of protective immunity against Plasmodium falciparum malaria. J Immunol (2010) 185(10):6157–67. doi:10.4049/jimmunol.1001555
99. Reiling L, Richards JS, Fowkes FJ, Wilson DW, Chokejindachai W, Barry AE, et al. The Plasmodium falciparum erythrocyte invasion ligand Pfrh4 as a target of functional and protective human antibodies against malaria. PLoS One (2012) 7(9):e45253. doi:10.1371/journal.pone.0045253
100. Bustamante LY, Bartholdson SJ, Crosnier C, Campos MG, Wanaguru M, Nguon C, et al. A full-length recombinant Plasmodium falciparum PfRH5 protein induces inhibitory antibodies that are effective across common PfRH5 genetic variants. Vaccine (2013) 31(2):373–9. doi:10.1016/j.vaccine.2012.10.106
101. Da DF, Dixit S, Sattabonkot J, Mu J, Abate L, Ramineni B, et al. Anti-Pfs25 human plasma reduces transmission of Plasmodium falciparum isolates that have diverse genetic backgrounds. Infect Immun (2013) 81(6):1984–9. doi:10.1128/IAI.00016-13
102. Hommel M, Elliott SR, Soma V, Kelly G, Fowkes FJ, Chesson JM, et al. Evaluation of the antigenic diversity of placenta-binding Plasmodium falciparum variants and the antibody repertoire among pregnant women. Infect Immun (2010) 78(5):1963–78. doi:10.1128/IAI.01365-09
103. Talundzic E, Shah S, Fawole O, Owino S, Moore JM, Peterson DS. Sequence polymorphism, segmental recombination and toggling amino acid residues within the DBL3X domain of the VAR2CSA placental malaria antigen. PLoS One (2012) 7(2):e31565. doi:10.1371/journal.pone.0031565
104. Gnidehou S, Jessen L, Gangnard S, Ermont C, Triqui C, Quiviger M, et al. Insight into antigenic diversity of VAR2CSA-DBL5epsilon domain from multiple Plasmodium falciparum placental isolates. PLoS One (2010) 5(10):e13105. doi:10.1371/journal.pone.0013105
105. Chen DS, Barry AE, Leliwa-Sytek A, Smith TA, Peterson I, Brown SM, et al. A molecular epidemiological study of var gene diversity to characterize the reservoir of Plasmodium falciparum in humans in Africa. PLoS One (2011) 6(2):e16629. doi:10.1371/journal.pone.0016629
106. Tanabe K, Arisue N, Palacpac NM, Yagi M, Tougan T, Honma H, et al. Geographic differentiation of polymorphism in the Plasmodium falciparum malaria vaccine candidate gene SERA5. Vaccine (2012) 30(9):1583–93. doi:10.1016/j.vaccine.2011.12.124
107. Patil A, Orjuela-Sanchez P, da Silva-Nunes M, Ferreira MU. Evolutionary dynamics of the immunodominant repeats of the Plasmodium vivax malaria-vaccine candidate circumsporozoite protein (CSP). Infect Genet Evol (2010) 10(2):298–303. doi:10.1016/j.meegid.2010.01.006
108. Zakeri S, Abouie Mehrizi A, Djadid ND, Snounou G. Circumsporozoite protein gene diversity among temperate and tropical Plasmodium vivax isolates from Iran. Trop Med Int Health (2006) 11(5):729–37. doi:10.1111/j.1365-3156.2006.01613.x
109. Dias S, Wickramarachchi T, Sahabandu I, Escalante AA, Udagama PV. Population genetic structure of the Plasmodium vivax circumsporozoite protein (Pvcsp) in Sri Lanka. Gene (2013) 518(2):381–7. doi:10.1016/j.gene.2013.01.003
110. Nobrega de Sousa T, Carvalho LH, Alves de Brito CF. Worldwide genetic variability of the Duffy binding protein: insights into Plasmodium vivax vaccine development. PLoS One (2011) 6(8):e22944. doi:10.1371/journal.pone.0022944
111. Chenet SM, Tapia LL, Escalante AA, Durand S, Lucas C, Bacon DJ. Genetic diversity and population structure of genes encoding vaccine candidate antigens of Plasmodium vivax. Malar J (2012) 11(1):68. doi:10.1186/1475-2875-11-68
112. Valizadeh V, Zakeri S, Mehrizi AA, Djadid ND. Population genetics and natural selection in the gene encoding the Duffy binding protein II in Iranian Plasmodium vivax wild isolates. Infect Genet Evol (2014) 21:424–35. doi:10.1016/j.meegid.2013.12.012
113. Ju HL, Kang JM, Moon SU, Bahk YY, Cho PY, Sohn WM, et al. Genetic diversity and natural selection of Duffy binding protein of Plasmodium vivax Korean isolates. Acta Trop (2013) 125(1):67–74. doi:10.1016/j.actatropica.2012.09.016
114. Kano FS, Sanchez BA, Sousa TN, Tang ML, Saliba J, Oliveira FM, et al. Plasmodium vivax Duffy binding protein: baseline antibody responses and parasite polymorphisms in a well-consolidated settlement of the Amazon region. Trop Med Int Health (2012) 17(8):989–1000. doi:10.1111/j.1365-3156.2012.03016.x
115. Ju HL, Kang JM, Moon SU, Kim JY, Lee HW, Lin K, et al. Genetic polymorphism and natural selection of Duffy binding protein of Plasmodium vivax Myanmar isolates. Malar J (2012) 11:60. doi:10.1186/1475-2875-11-60
116. Premaratne PH, Aravinda BR, Escalante AA, Udagama PV. Genetic diversity of Plasmodium vivax Duffy binding protein II (PvDBPII) under unstable transmission and low intensity malaria in Sri Lanka. Infect Genet Evol (2011) 11(6):1327–39. doi:10.1016/j.meegid.2011.04.023
117. Collins WE, Jeffery GM, Roberts JM. A retrospective examination of reinfection of humans with Plasmodium vivax. Am J Trop Med Hyg (2004) 70(6):642–4.
118. Jeffery GM. Epidemiological significance of repeated infections with homologous and heterologous strains and species of Plasmodium. Bull World Health Organ (1966) 35(6):873–82.
119. McKenzie FE, Smith DL, O’Meara WP, Riley EM. Strain theory of malaria: the first 50 years. Adv Parasitol (2008) 66:1–46. doi:10.1016/S0065-308X(08)00201-7
120. Bull PC, Lowe BS, Kortok M, Molyneux CS, Newbold CI, Marsh K. Parasite antigens on the infected red cell surface are targets for naturally acquired immunity to malaria. Nat Med (1998) 4(3):358–60. doi:10.1038/nm0398-358
121. Day KP, Marsh K. Naturally acquired immunity to Plasmodium falciparum. Immunol Today (1991) 12(3):A68–71. doi:10.1016/S0167-5699(05)80020-9
122. Bai T, Becker M, Gupta A, Strike P, Murphy VJ, Anders RF, et al. Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket. Proc Natl Acad Sci U S A (2005) 102(36):12736–41. doi:10.1073/pnas.0501808102
123. Collins CR, Withers-Martinez C, Hackett F, Blackman MJ. An inhibitory antibody blocks interactions between components of the malarial invasion machinery. PLoS Pathog (2009) 5(1):e1000273. doi:10.1371/journal.ppat.1000273
124. Nielsen MA, Pinto VV, Resende M, Dahlback M, Ditlev SB, Theander TG, et al. Induction of adhesion-inhibitory antibodies against placental Plasmodium falciparum parasites by using single domains of VAR2CSA. Infect Immun (2009) 77(6):2482–7. doi:10.1128/IAI.00159-09
125. Ntumngia FB, King CL, Adams JH. Finding the sweet spots of inhibition: understanding the targets of a functional antibody against Plasmodium vivax Duffy binding protein. Int J Parasitol (2012) 42(12):1055–62. doi:10.1016/j.ijpara.2012.09.006
126. Dutta S, Lee SY, Batchelor AH, Lanar DE. Structural basis of antigenic escape of a malaria vaccine candidate. Proc Natl Acad Sci U S A (2007) 104(30):12488–93. doi:10.1073/pnas.0701464104
127. Takala SL, Coulibaly D, Thera MA, Batchelor AH, Cummings MP, Escalante AA, et al. Extreme polymorphism in a vaccine antigen and risk of clinical malaria: implications for vaccine development. Sci Transl Med (2009) 1(2):2ra5. doi:10.1126/scitranslmed.3000257
128. Reeder JC, Wapling J, Mueller I, Siba PM, Barry AE. Population genetic analysis of the Plasmodium falciparum 6-cys protein Pf38 in Papua New Guinea reveals domain-specific balancing selection. Malar J (2011) 10:126. doi:10.1186/1475-2875-10-126
129. Arnott A, Mueller I, Ramsland PA, Siba PM, Reeder JC, Barry AE. Global population structure of the genes encoding the malaria vaccine candidate, Plasmodium vivax apical membrane antigen 1 (PvAMA1). PLoS Negl Trop Dis (2013) 7(10):e2506. doi:10.1371/journal.pntd.0002506
130. Taylor RR, Smith DB, Robinson VJ, McBride JS, Riley EM. Human antibody response to Plasmodium falciparum merozoite surface protein 2 is serogroup specific and predominantly of the immunoglobulin G3 subclass. Infect Immun (1995) 63(11):4382–8.
131. Ouattara A, Takala-Harrison S, Thera MA, Coulibaly D, Niangaly A, Saye R, et al. Molecular basis of allele-specific efficacy of a blood-stage malaria vaccine: vaccine development implications. J Infect Dis (2013) 207(3):511–9. doi:10.1093/infdis/jis709
132. Kusi KA, Faber BW, Thomas AW, Remarque EJ. Humoral immune response to mixed PfAMA1 alleles; multivalent PfAMA1 vaccines induce broad specificity. PLoS One (2009) 4(12):e8110. doi:10.1371/journal.pone.0008110
133. Remarque EJ, Faber BW, Kocken CH, Thomas AW. A diversity- covering approach to immunization with Plasmodium falciparum apical membrane antigen 1 induces broader allelic recognition and growth inhibition responses in rabbits. Infect Immun (2008) 76(6):2660–70. doi:10.1128/IAI.00170-08
134. Faber BW, Younis S, Remarque EJ, Rodriguez Garcia R, Riasat V, Walraven V, et al. Diversity covering AMA1-MSP119 fusion proteins as malaria vaccines. Infect Immun (2013) 81(5):1479–90. doi:10.1128/IAI.01267-12
135. Coley AM, Parisi K, Masciantonio R, Hoeck J, Casey JL, Murphy VJ, et al. The most polymorphic residue on Plasmodium falciparum apical membrane antigen 1 determines binding of an invasion-inhibitory antibody. Infect Immun (2006) 74(5):2628–36. doi:10.1128/IAI.74.5.2628-2636.2006
136. Conway DJ, Cavanagh DR, Tanabe K, Roper C, Mikes ZS, Sakihama N, et al. A principal target of human immunity to malaria identified by molecular population genetic and immunological analyses. Nat Med (2000) 6(6):689–92. doi:10.1038/76272
137. Weedall GD, Conway DJ. Detecting signatures of balancing selection to identify targets of anti-parasite immunity. Trends Parasitol (2010) 26(7):363–9. doi:10.1016/j.pt.2010.04.002
138. Takala SL, Coulibaly D, Thera MA, Dicko A, Smith DL, Guindo AB, et al. Dynamics of polymorphism in a malaria vaccine antigen at a vaccine-testing site in Mali. PLoS Med (2007) 4(3):e93. doi:10.1371/journal.pmed.0040093
139. Anderson TJ, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, et al. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol (2000) 17(10):1467–82. doi:10.1093/oxfordjournals.molbev.a026247
140. Imwong M, Nair S, Pukrittayakamee S, Sudimack D, Williams JT, Mayxay M, et al. Contrasting genetic structure in Plasmodium vivax populations from Asia and South America. Int J Parasitol (2007) 37(8–9):1013–22. doi:10.1016/j.ijpara.2007.02.010
141. Koepfli C, Mueller I, Marfurt J, Goroti M, Sie A, Oa O, et al. Evaluation of Plasmodium vivax genotyping markers for molecular monitoring in clinical trials. J Infect Dis (2009) 199(7):1074–80. doi:10.1086/597303
142. Falk N, Maire N, Sama W, Owusu-Agyei S, Smith T, Beck HP, et al. Comparison of PCR-RFLP and Genescan-based genotyping for analyzing infection dynamics of Plasmodium falciparum. Am J Trop Med Hyg (2006) 74(6):944–50.
143. Betuela I, Rosanas-Urgell A, Kiniboro B, Stanisic DI, Samol L, de Lazzari E, et al. Relapses contribute significantly to the risk of Plasmodium vivax infection and disease in Papua New Guinean children 1-5 years of age. J Infect Dis (2012) 206(11):1771–80. doi:10.1093/infdis/jis580
144. Barry AE, Leliwa A, Choi M, Nielsen KM, Hartl DL, Day KP. DNA sequence artifacts and the estimation of time to the most recent common ancestor (TMRCA) of Plasmodium falciparum. Mol Biochem Parasitol (2003) 130(2):143–7. doi:10.1016/S0166-6851(03)00164-6
145. Chan ER, Menard D, David PH, Ratsimbasoa A, Kim S, Chim P, et al. Whole genome sequencing of field isolates provides robust characterization of genetic diversity in Plasmodium vivax. PLoS Negl Trop Dis (2012) 6(9):e1811. doi:10.1371/journal.pntd.0001811
146. Duan J, Mu J, Thera MA, Joy D, Kosakovsky Pond SL, Diemert D, et al. Population structure of the genes encoding the polymorphic Plasmodium falciparum apical membrane antigen 1: implications for vaccine design. Proc Natl Acad Sci U S A (2008) 105(22):7857–62. doi:10.1073/pnas.0802328105
147. Excoffier L, Heckel G. Computer programs for population genetics data analysis: a survival guide. Nat Rev Genet (2006) 7(10):745–58. doi:10.1038/nrg1904
148. Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics (1989) 123(3):585–95.
149. Parobek CM, Bailey JA, Hathaway NJ, Socheat D, Rogers WO, Juliano JJ. Differing patterns of selection and geospatial genetic diversity within two leading Plasmodium vivax candidate vaccine antigens. PLoS Negl Trop Dis (2014) 8(4):e2796. doi:10.1371/journal.pntd.0002796
150. Bruce MC, Galinski MR, Barnwell JW, Donnelly CA, Walmsley M, Alpers MP, et al. Genetic diversity and dynamics of Plasmodium falciparum and P. vivax populations in multiply infected children with asymptomatic malaria infections in Papua New Guinea. Parasitology (2000) 121(Pt 3):257–72. doi:10.1017/S0031182099006356
151. Stanisic DI, Javati S, Kiniboro B, Lin E, Jiang J, Singh B, et al. Naturally acquired immune responses to P. vivax merozoite surface protein 3alpha and merozoite surface protein 9 are associated with reduced risk of P. vivax malaria in young Papua New Guinean children. PLoS Negl Trop Dis (2013) 7(11):e2498. doi:10.1371/journal.pntd.0002498
152. Barnadas C, Kent D, Timinao L, Iga J, Gray LR, Siba P, et al. A new high-throughput method for simultaneous detection of drug resistance associated mutations in Plasmodium vivax dhfr, dhps and mdr1 genes. Malar J (2011) 10:282. doi:10.1186/1475-2875-10-282
Keywords: Plasmodium falciparum, Plasmodium vivax, malaria, vaccine, strain, diversity, polymorphism, clinical trials
Citation: Barry AE and Arnott A (2014) Strategies for designing and monitoring malaria vaccines targeting diverse antigens. Front. Immunol. 5:359. doi: 10.3389/fimmu.2014.00359
Received: 12 May 2014; Accepted: 13 July 2014;
Published online: 28 July 2014.
Edited by:
Magdalena Plebanski, Monash University, AustraliaReviewed by:
Hajime Hisaeda, Gunma University, JapanShannon Takala Harrison, University of Maryland School of Medicine, USA
Copyright: © 2014 Barry and Arnott. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alyssa E. Barry, Division of Infection and Immunity, Walter and Eliza Hall Institute for Medical Research, 1G Royal Parade, Parkville, VIC 3052, Australia e-mail: barry@wehi.edu.au
 Alyssa E. Barry
Alyssa E. Barry Alicia Arnott
Alicia Arnott