Transcriptomic Abnormalities in Epstein Barr Virus Associated T/NK Lymphoproliferative Disorders
- 1Department of Haematology-Oncology, National University Cancer Institute of Singapore, National University Health System, Singapore, Singapore
- 2Department of Medicine, National University Health System, Singapore, Singapore
- 3Cancer Science Institute of Singapore, National University of Singapore, Singapore, Singapore
- 4Department of Pathology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore
- 5Department of Pathology, National University Health System, Singapore, Singapore
Epstein Barr virus positive T/NK lymphoproliferative disorders (EBV-TNKLPD) comprise a spectrum of neoplasms ranging from cutaneous lymphoid proliferations to aggressive lymphomas. The spectrum includes extranodal NK/T-cell lymphoma (ENKTL), aggressive NK-cell leukemia, and a group of EBV-TNKLPDs affecting children which are poorly characterized in terms of their molecular biology. Gene and miRNA expression profiling has elucidated RNA abnormalities which impact on disease biology, classification, and treatment of EBV-TNKLPD. Pathways promoting proliferation, such as Janus associated kinase/ Signal Transducer and Activator of Transcription (JAK/STAT) and nuclear factor kB, are upregulated in ENKTL while upregulation of survivin and deregulation of p53 inhibit apoptosis in both ENKTL and chronic active EBV infection (CAEBV). Importantly, immune evasion via the programmed cell death-1 and its ligand, PD-1/PD-L1 checkpoint pathway, has been demonstrated to play an important role in ENKTL. Other pathogenic mechanisms involve EBV genes, microRNA deregulation, and a variety of other oncogenic signaling pathways. The identification of EBV-positive Peripheral T-cell lymphoma not otherwise specified (PTCL-NOS) as a tumor with a distinct molecular signature and clinical characteristics highlights the important contribution of the knowledge derived from gene and miRNA expression profiling in disease classification. Novel therapeutic targets identified through the study of RNA abnormalities provide hope for patients with EBV-TNKLPD, which often has a poor prognosis. Immune checkpoint inhibition and JAK inhibition in particular have shown promise and are being evaluated in clinical trials. In this review, we provide an overview of the key transcriptomic aberrancies in EBV-TNKLPD and discuss their translational potential.
Introduction
Epstein–Barr virus (EBV) is a ubiquitous human herpesvirus with B-cell tropism and the ability to transform infected B lymphocytes into continuously proliferating lymphoblastoid cells. Infrequently, EBV infects T cells and Natural Killer (NK) cells, which can result in a wide spectrum of EBV-positive cytotoxic T/NK cell lymphoproliferative diseases (EBV-TNKLPD). The current classification of EBV-TNKLPD includes (i) systemic chronic active EBV infection of T- and NK-cell type (CAEBV), (ii) cutaneous CAEBV, which includes hydroa vacciniforme-like lymphoproliferative disorder (HV-LPD) and severe mosquito bite allergy (MBA), (iii) systemic EBV-positive T-cell lymphoma of childhood (STCL), (iv) aggressive NK-cell leukemia (ANKL), (v) extranodal NK/T-cell lymphoma, nasal-type (ENKTL), and (vi) nodal peripheral T-cell lymphoma, EBV-positive (EBV-PTCL) (1–3). ENKTL and ANKL are the prototypic examples of EBV-TNKLPD and are well-recognized (4). On the other hand, the classification of EBV-TNKLPD occurring in childhood (EBV-TNKLPD-childhood) has evolved and was recently updated in the revised 4th edition of the WHO classification (1). It includes a spectrum of diseases with heterogeneous clinical manifestations, a broad range of morphology from polymorphic to monomorphic lymphoid proliferations, and indolent behavior to systemic and aggressive diseases (5). EBV-TNKLPD involving primarily lymph nodes is uncommon but shows characteristic clinical and molecular features distinct from ENKTL (6). This group of nodal EBV-TNKLPD is currently classified as an EBV-positive variant of PTCL, not otherwise specified (EBV-PTCL), as it is presently unclear whether they represent a distinct entity (2).
The spectrum of EBV-TNKLPD remains a challenging group of diseases to study and this is often attributed to the rarity of disease and limited tissue availability (7, 8). Due to significant overlap in morphology and phenotype, the precise distinction of each of the entities can be challenging (9). Furthermore, the nomenclature and classification of EBV-TNKLPD occurring in childhood has been confusing and has suffered from a lack of well-established diagnostic criteria until recently (3, 10).
Genome-wide gene expression profiling (GEP) has revolutionized and greatly improved our understanding of the molecular biology of lymphoma (11, 12). Similarly, GEP has identified robust molecular signatures for subtypes of PTCL and ENKTL and uncovered actionable therapeutic targets (13, 14). Novel insights gleaned from recent genome-wide high throughout techniques have also significantly advanced our understanding of ENKTL (8, 15–18). On the other hand, knowledge of the molecular biology and genomics of EBV-TNKLPD of childhood (EBV-TNKLPD-childhood), such as CAEBV, is only slowly unraveling and there remains a paucity of large-scale gene or miRNA profiling of the diseases in childhood. In this brief review, we will summarize current insights on the transcriptomics abnormalities in EBV-TNKLPD and focus on those with translational impact on (i) understanding molecular biology and disease pathogenesis, (ii) disease classification and refining diagnosis, (iii) disease prognosis, and (iv) therapy. An overview of these abnormalities and their role in the pathogenesis of EBV-TNKLPD is presented in Figure 1.
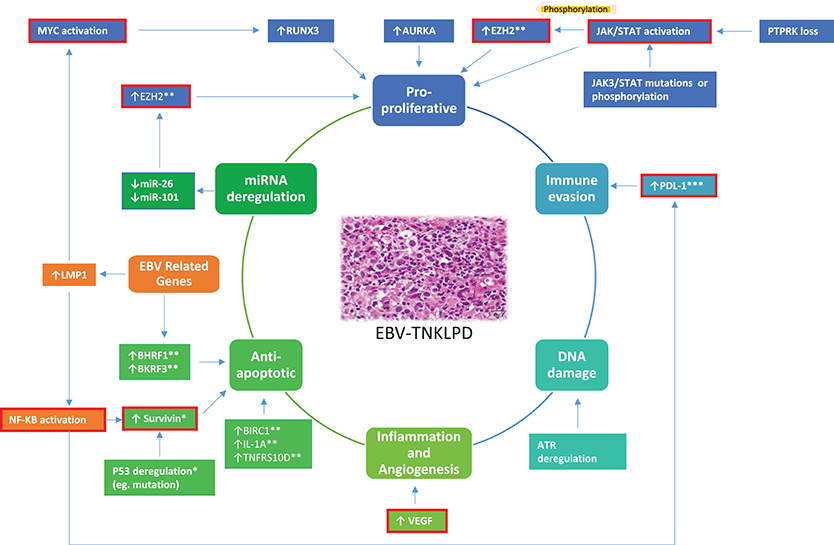
Figure 1. A proposed model highlighting an overview of the main RNA abnormalities in EBV-TNKLPD. The key processes involved in the lymphomagenesis of EBV-TNKLPD are indicated in the central circle of the figure. The RNA abnormalities are grouped and color-coded according to the postulated processes and those which are likely to contribute to multiple processes are highlighted in orange. RNA abnormalities which have potential translational impact are outlined with red. All the above RNA abnormalities are present in ENKTL. *Refers to RNA abnormalities which are present in ENKTL, Systemic EBV+ T-Cell Lymphoma and CAEBV. **Refers to RNA abnormalities which are present in ENKTL and CAEBV. ***Refers to RNA abnormalities which are present in ENKTL and EBV+ PTCL NOS.
Impact of Transcriptomic Abnormalities on Understanding Disease Pathogenesis or Deregulated Pathways
Cellular Proliferation
Janus Kinase/Signal Transducer and Activator of Transcription (JAK/STAT) signaling has recently been shown to play a prominent role in the pathogenesis of ENKTL and ANKL (19, 20). GEP studies have revealed that components of this pathway are differentially expressed in ENKTL compared to normal NK cells (15, 21). Transcriptomic sequencing and integrated genomic analysis of ANKL showed that JAK/STAT mutations resulted in overexpression of MYC and its interacting proteins (20). These data suggest that JAK/STAT signaling promotes proliferation of malignant NK cells not only through its known pro-proliferative function but also through interaction with oncogenes, such as MYC. MYC is upregulated in ENKTL, and this is associated with a corresponding overexpression of its transcriptional targets. (17). Among these MYC targets is RUNX3, which promotes proliferation and survival in ENTKL (22). Inhibition of MYC leads to downregulation of RUNX3 and apoptosis in ENTKL cells, which supports a potential therapeutic role of MYC in ENKTL (22).
Enhancer of Zeste Homolog 2 (EZH2), a component of the polycomb repressive complex 2 (PRC2), has also been shown to be upregulated in ENKTL (17, 23). This may be explained by Myc-induced downregulation of microRNAs, miR-26a, and miR-101, which negatively regulate EZH2 expression (23). In ENKTL, EZH2 does not function as an epigenetic regulator. Instead, it acts as a transcriptional co-activator via a non-canonical pathway (24), and this switch of function is mediated by JAK3 via phosphorylation of EZH2. Inhibition of JAK3 has been demonstrated to reduce the proliferation of ENKTL cells, indicating that targeting this pathway may be a potential therapeutic strategy (25). EZH2 has also been shown to be upregulated in EBV-TNKLPD-childhood and, similar to ENKTL, downregulation of EZH2 using a PRC2 inhibitor induces apoptosis in CAEBV cell lines (23, 26).
Nuclear factor (NF) kB is a transcription factor with pro-survival and anti-apoptotic functions known to be upregulated in lymphoid malignancies (27). NF-kB and its target genes were shown to be overexpressed in ENKTL by GEP in two studies (15, 17) and treatment of ENKTL cell lines with NF-kB inhibitors resulted in induction of apoptosis (28). However, these results were not replicated in another study comparing ENKTL to PTCL (16). Aurora kinase A (AURKA) is a mitotic kinase important for cell proliferation. AURKA is also overexpressed in ENKTL and targeted inhibition induced significant growth arrest in ENKTL cell lines (16, 17). Recent data suggest that AURKA interacts with MYC and WNT pathways to promote cell proliferation (29, 30). Further studies are required to better delineate the role of AURKA in ENKTL pathogenesis.
Murakami et al. performed GEP on CAEBV and ENKTL samples and identified upregulation of interleukin-2 (IL-2), IL-10, interferon gamma receptor 1 (IFNGR1), and Inhibin beta A (INHBA) (31). It has been proposed that EBV-infected T cells secrete IL-2 which functions via an autocrine loop to promote proliferation of T-cells in CAEBV. The binding of IFN-γ to IFNGR1 leads to activation of the JAK-STAT pathway, which is amenable to targeted therapy. (32, 33) The precise role of INHBA and IL-10 in EBV-TNKLPD remains unknown (31). Other genes that may confer a proliferative signature and are upregulated in ENKTL and CAEBV cell lines include cyclin dependent kinase 2 (CDK2), a regulator of cell cycle progression, and heat shock 90kDa protein 1-alpha (HSPCA), which is important for normal protein folding and survival of cancer cells (34).
In summary, there are multiple pathways which may provide mitogenic signals and allow the neoplastic cells to survive and proliferate in ENKTL and CAEBV. Among the pathways described above, JAK/STAT and NF-kB are the best studied in ENKTL. EZH2 and AURKA have promising translational impact and may serve as potential therapeutic targets in ENKTL. The evaluation of JAK3 inhibitors as modulators of non-canonical EZH2 activity in clinical trials is warranted.
Deregulation of Apoptosis and the DNA Damage Response
Resistance to apoptosis is a known hallmark of cancer and development of drug resistance (35). Several protein families that act as negative regulators of apoptosis by inhibiting cell death signaling pathways have been reported to be upregulated in CAEBV and ENKTL, including BIRC1, interleukin 1 alpha (IL1A), tumor necrosis factor receptor superfamily member 10d (TNFRS10D), survivin, p53, and NF-kB (15, 17, 26, 34, 36). Survivin is an anti-apoptotic protein that is overexpressed in the majority of ENKTL and EBV-TNKLPD-childhood (17, 26). Treatment of ENKTL cells with a survivin inhibitor, terameprocol, leads to increased apoptosis, suggesting this may be a therapeutic target. p53 is upregulated in ENKTL and EBV-TNKLPD-childhood and this is associated with deregulation of genes which are normally controlled by p53 (17, 26). p53 deregulation in ENKTL may be caused by mutations as reported by Quintanilla-Martinez et al. (36).
Appropriate response to DNA damage is essential for the maintenance of genome stability. Ataxia-telangiectasia mutated (ATM) and ataxia-telangiectasia-related (ATR) kinases are central regulators of the DNA damage response signaling pathway (37). Deregulation of ATR protein has been reported in ENKTL and CAEBV due to deletions resulting from aberrant splicing and leads to an abnormal response to DNA damage (38). ATR and related cell cycle checkpoint genes were also found to be overexpressed in ENKTL in another study by Ng et al. (17).
These data suggest that a defective DNA damage response along with inhibition of apoptosis may contribute to lymphomagenesis of EBV-TNKLPD. Survivin and p53 stand out as key players in the pathogenesis of EBV-TNKLPD. Survivin in particular may have potential as a therapeutic target based on the pre-clinical data discussed. Non-apoptotic cell death pathways are also a potential research arena for drug discovery and targeted therapies and are especially important for the circumvention of drug resistance (35).
Immune Evasion
Programmed cell death-1 (PD-1) inhibition has changed the landscape of immunotherapy for lymphoid malignancies (39, 40). GEP studies have shown that PD-Ligand 1 (PD-L1, also known as CD274) mRNA is upregulated in ENTKL compared to control tissues (6, 8). Overexpression of PD-L1 in ENKTL has been proposed to be mediated by LMP1 via MAPK, NF-kB, and STAT3 signaling (41, 42). The glycoprotein, CD38, is expressed in the majority of ENKTL and its expression is associated with an inferior outcome (8, 43). We have demonstrated through GEP study that CD38 is upregulated in ENKTL compared to control tissues (GEO database GSE90597) (8). Recent in vitro studies revealed that daratumumab, a humanized monoclonal antibody approved for the treatment of relapsed multiple myeloma, has good efficacy against ENKTL (44). Our current understanding of the role of PD1 and CD38 in EBV-TNKLPD remains incomplete. Novel regulators of PD1 such as CMTM6 (45) warrant investigation in this context while the function of CD38 in lymphomagenesis requires further study.
Whole-transcriptome microarray studies have identified a unique set of 30 genes which are dysregulated in CAEBV (46). These include several phagocytosis-associated genes such as C1QC, FGL232, and PSTPIP233 as well as monocyte markers FCGR1A and FCGR1B (CD64A/B), suggesting a relatively hyperactive phagocytosis and monocyte-mediated antibody-dependent cellular cytotoxicity in CAEBV (46). The expression of many CAEBV-unique genes was highly correlated with the level of CD64, indicating an important role for monocytes in the cellular immune response to CAEBV (46). Understanding the immune microenvironment of EBV-TNKLPD will be helpful in the incorporation of immunotherapy in this group of diseases. The PD-1/PD-L1 pathway is the most important transcriptomic abnormality from a biological and translational point of view. The potential of this pathway as a therapeutic target is discussed below.
Tumor Promoting Inflammation and Angiogenesis
Chronic inflammation is a known driver of malignancy and angiogenesis is critical for tumor growth and metastasis (47). Vascular endothelial growth factor (VEGF) promotes tumor vascularization and growth in a variety of malignancies (48). VEGF is upregulated in ENKTL and has been proposed as a therapeutic target (7, 49). Guanylate-binding protein 1 (GBP1), a G protein involved in the chronic inflammatory response and strongly induced in endothelial cells and lymphocytes, was found to be overexpressed in CAEBV cells (50). It is postulated that the upregulation of IFNGR1 in CAEBV may result in the overexpression of GBP1, which in turn contributes to vascular dysfunction in chronic inflammation (31). Tumor necrosis factor alpha-induced protein 6 (TNFAIP6) is an adhesion molecule that plays multiple roles in chronic inflammation and tissue remodeling. TNFAIP6 is upregulated in CAEBV and postulated to play a similar role to GBP1 in this context (50). Activated T-cells in CAEBV express higher levels of interleukin-10 (IL-10), transforming growth factor-β (TGF-β), and IFN-γ (51), with the expression of IL-10 and TGF-β being proportional to the EBV viral load in T cells (51). These data suggest that a complex deregulation of pro-inflammatory cytokines driven by EBV as well as a potent angiogenic drive play a crucial role in the pathogenesis of EBV-TNKLPD. VEGF appears to have the greatest translational potential among the deregulated angiogenic pathways discussed and requires further study.
EBV Related Genes
EBV mediated oncogenesis is thought to be driven by genes expressed during latency, such as LMP1 (52). The expression of EBV-related lytic genes, such as BHRF1 and BKRF3, was found to be increased in ENKTL cell lines and may have an anti-apoptotic role as BHRF1 has sequence homology with human BCL-2 (34). BZLF1, which encodes the immediate-early gene product Zta, was preferentially expressed in CAEBV compared to ENKTL cell lines (34). Given the critical role of EBV, further studies are required to fully understand the mechanistic underpinnings of the virus in the lymphomagenesis of this spectrum of disease with the aim of developing therapeutic targets.
Other Oncogenic Signaling Pathways
Other signaling pathways reported to play a pathogenic role in ENKTL include PDGFRα, AKT, and NOTCH-1 (7, 15). The availability of inhibitors to the NOTCH and AKT signaling pathways is currently under evaluation, and their clinical efficacy in ENKTL remains to be established (53, 54).
MicroRNA Deregulation
MicroRNAs (miRNA) have a critical role in the regulation of gene expression in cancer and have been proposed to play an important role in ENKTL and CAEBV (55, 56). miRNAs in ENKTL are predominantly downregulated compared to normal NK cells, specific examples include miR-150, miR-101, miR-26a, miR-26b, miR-28-5, miR-363, and miR-146 (57, 58). The targets of these miRNAs include genes in critical pathways such as p53, MAPK, and EZH2 (23, 57). Less commonly, miRNAs with pro-oncogenic functions, such as miR-21 and miR-155, are upregulated in ENKTL (59). miRNA profiles have also been suggested to have prognostic significance. For instance, downregulation of miR-146a and upregulation of miR221 are associated with poor prognosis (60, 61). The utility of miRNAs as a therapeutic target is, however, unknown and requires further studies.
Less is known about the miRNA profile of CAEBV than ENKTL. In a study of patients with CAEBV compared to those with infectious mononucleosis and healthy controls, miR-BART 1-5p, 2-5p, 5, and 22 were found to be upregulated in CAEBV patients (62). Interestingly, miR-BART 13, miR-BART 2-5p, and 15 levels were higher in patients with active compared to inactive disease, suggestive of a potential role in monitoring disease activity (56, 62).
Impact of Transcriptomic Abnormalities on Disease Classification and Refining Diagnosis
In addition to characterizing deregulated oncogenic pathways, GEP studies have provided new perspectives on the molecular biology, ontogeny, and classification of ENKTL. The GEP of ENKTL is distinct from PTCL NOS and shows higher expression of genes associated with NK cell lineage (14, 15). In addition, ENKTL demonstrated a similar molecular signature to a subset of gamma-delta (γδ) T-cell lymphomas that are non-hepatosplenic in presentation and akin to a subset of ENKTL derived from γδ T-cells (16). Interestingly, these “molecularly defined” γδ T-cell lymphomas had similar clinical outcomes to ENKTL (14, 16).
EBV-PTCL is an EBV-associated T/NK cell lymphoma with primary nodal disease presentation and shows molecular and clinical features distinct from ENKTL, including older age, lack of nasal involvement and CD8-positive/CD56-negative phenotype (6). Gene set enrichment analysis revealed significant enrichment for hallmark gene sets, such as MTORC1_SIGNALING, IL6_JAK_STAT3_SIGNALING as well as several gene sets related to cell cycle and genomic instability, including G2M_CHECKPOINT, E2F_TARGETS, MYC_TARGETS, and APOPTOSIS. Current data support the WHO proposal to classify this disease separately from ENKTL and this distinction is clinically important as EBV-PTCL is significantly more aggressive than ENKTL and should be managed differently (6).
Since there is significant overlap in the clinicopathologic features of ENKTL and EBV-TNKLPD-childhood, studies have also been conducted to compare the molecular signature of these entities. GEP results indicated a high degree of similarity between EBV-TNKLPD-childhood and ENKTL, with overexpression of p53, survivin, and EZH2 (26). Notably, there is a distinctive enrichment of stem cell-related genes in EBV-TNKLPD-childhood compared to ENKTL (26). The discovery of potential cancer stem cell phenotype in EBV-TNKLPD-childhood has potential therapeutic implications and may explain why conventional chemotherapy, without hematopoietic stem cell transplant, is often unsuccessful in the treatment of the disease in childhood (63, 64).
Impact of Transcriptomic Abnormalities on Disease Prognostication
The challenge in EBV-TNKLPD-childhood remains to identify morphological or clinical markers to predict outcome. While age, liver dysfunction, and treatment with transplantation play a role in prognosis (65), criteria such as presence of systemic symptoms, T-cell clonality, amount of EBV-positive cells, and/or density of the infiltrate do not help in predicting disease progression (66). In this regard, Ng et al. compared the molecular signature between 2 groups of EBV-TNKLPD-childhood with different outcomes and identified overexpression of cyclinE2 gene and protein to be significantly associated with poor outcome (67).
The prognostication of ENKTL is largely based on clinical features (68). Transcriptomics abnormalities have not made a significant contribution to the risk stratification of ENKTL although there is a suggestion that specific miRNAs (miR-146a and miR-221) may have a prognostic impact (60, 61).
Impact of Transcriptomic Abnormalities on Therapy
Transcriptomics aberrancies in EBV-TNKLPD have provided insight into potential therapeutic targets. Among the most promising is immune checkpoint inhibition. Overexpression of PD-L1 mRNA and protein has been demonstrated in ENKTL and EBV-PTCL (6, 8) and patients with relapsed ENKTL have shown remarkable response to pembrolizumab, an antibody against PD1 (69). However, there is a lack of correlation between clinical response to pembrolizumab with PD-L1 expression in tumor cells. Further research is therefore required to understand the mechanism of action and to identify new predictive biomarkers for checkpoint inhibition.
The anti-CD38 monoclonal antibody, daratumumab, is approved for multiple myeloma (70). Recent in vitro studies revealed that daratumumab may also have efficacy against ENKTL (44). This hypothesis was supported clinically by the dramatic response of a patient with refractory ENKTL to daratumumab monotherapy (71). Targeting LMP1/2 using cytotoxic T-lymphocytes has also revealed encouraging results in EBV-associated lymphomas. While these data have shown initial promise, further study is required to evaluate the mechanism of action and clinical efficacy of these agents (40).
The JAK-STAT pathway is another potentially useful therapeutic target with JAK3 and STAT3 inhibitors showing in vitro activity against ENKTL (25, 72). JAK inhibitors are currently being evaluated against ENKTL in a phase 2 clinical trial (NCT02974647) (73). The potential for JAK inhibition to target the INF-γ pathway in CAEBV is also an attractive therapeutic option requiring further study (31). Proteasome inhibitors to target NF-kB have been evaluated in clinical trials in combination with chemotherapy and additional evaluation is necessary to assess their efficacy in ENKTL (74, 75). Other potential therapeutic targets based on preliminary in vitro data on ENKTL include PDGFRa, VEGF, AURKA, NOTCH, CDK2, MYC, EZH2, and survivin (7, 8, 15–17, 22, 23).
Conclusions and Future Directions
Understanding gene and miRNA transcriptomic abnormalities in EBV TNKLPD has improved our understanding of the molecular biology of this group of tumors, with an impact on disease classification, prognosis, and treatment. The key abnormalities associated with each entity are summarized in Table 1 and an overview of the major deregulated pathways is represented in Figure 1. Further characterization of the molecular signatures of these tumors, especially those occurring in childhood, will help direct functional studies on the disease pathogenesis and decipher the role of EBV while stimulating insights into development of new treatment strategies for these patients.
Author Contributions
S-BN and W-JC conceptualized the review. SdM and S-BN wrote the manuscript. JT prepared the figure and table. AJ and W-JC critically reviewed and edited the manuscript, figure, and table.
Funding
S-BN was supported by the National Medical Research Council, Clinician Scientist Award (WBS R–179–000–063–213).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Quintanilla-Martinez L, Ko YH, Kimura Jaffe HES. EBV-positive T-cell and NK-cell lymphoproliferative diseases of childhood. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: International Agency for Research on Cancer (2017) 355–363.
2. Chan JKC, Quintanilla-Martinez L, Ferry JA. Extranodal NK/T-cell lymphoma, nasal type. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: International Agency for Research on Cancer (2017) 368–371.
3. Ko YH, Chan JKC, Quintanilla-Martinez L. Virally associated T-cell and NK-cell Neoplasms. In: Jaffe ES, Arber DA, Campo EN, Harris L, Quintanilla-Martinez L, editors. Hematopathology. Philadelphia, PA: Elsevier (2017) 565–598.
4. Ng SB, Khoury JD. Epstein-Barr virus in lymphoproliferative processes: an update for the diagnostic pathologist. Adv Anat Pathol. (2009) 16:40–55. doi: 10.1097/PAP.0b013e3181916029
5. Ohshima K, Kimura H, Yoshino T, Kim CW, Ko YH, Lee SS, et al. Proposed categorization of pathological states of EBV-associated T/natural killer-cell lymphoproliferative disorder (LPD) in children and young adults: overlap with chronic active EBV infection and infantile fulminant EBV T-LPD. Pathol Int. (2008) 58:209–17. doi: 10.1111/j.1440-1827.2008.02213.x
6. Ng SB, Chung TH, Kato S, Nakamura S, Takahashi E, Ko YH, et al. Epstein-Barr virus-associated primary nodal T/NK-cell lymphoma shows a distinct molecular signature and copy number changes. Haematologica (2018) 103:278–87. doi: 10.3324/haematol.2017.180430
7. Huang Y, de Leval L, Gaulard P. Molecular underpinning of extranodal NK/T-cell lymphoma. Best Pract Res Clin Haematol. (2013) 26:57–74. doi: 10.1016/j.beha.2013.04.006
8. de Mel S, Soon GS, Mok Y, Chung TH, Jeyasekharan AD, Chng WJ, et al. The genomics and molecular biology of natural killer/T-Cell lymphoma: opportunities for translation. Int J Mol Sci. (2018) 19:1931. doi: 10.3390/ijms19071931
9. Attygalle D, Cabecadas J, Gaulard P, Jaffe ES, de Jong D, Ko Yv. Peripheral T-cell and NK-cell lymphomas and their mimics; taking a step forward - report on the lymphoma workshop of the XVIth meeting of the European Association for Haematopathology and the Society for Hematopathology. Histopathology (2014) 64:171–99. doi: 10.1111/his.12251
10. Ko YH, Kim HJ, Oh YH, Park G, Lee SS, Huh J, et al. EBV-associated T and NK cell lymphoproliferative disorders: consensus report of the 4th Asian Hematopathology Workshop. J Hematopathol. (2012) 5:319–24. doi: 10.1007/s12308-012-0169-1
11. Orsborne C, Byers R. Impact of gene expression profiling in lymphoma diagnosis and prognosis. Histopathology (2011) 58:106–27. doi: 10.1111/j.1365-2559.2010.03708.x
12. Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature (2000) 403:503–11. doi: 10.1038/35000501
13. Iqbal J, Wilcox R, Naushad H, Rohr J, Heavican TB, Wang C, et al. Genomic signatures in T-cell lymphoma: how can these improve precision in diagnosis and inform prognosis? Blood Rev. (2016) 30:89–100. doi: 10.1016/j.blre.2015.08.003
14. Iqbal J, Wright G, Wang C, Rosenwald A, Gascoyne RD, Weisenburger DD, et al. Gene expression signatures delineate biological and prognostic subgroups in peripheral T-cell lymphoma. Blood (2014) 123:2915–23. doi: 10.1182/blood-2013-11-536359
15. Huang Y, de Reynies A, de Leval L, Ghazi B, Martin-Garcia N, Travert M, et al. Gene expression profiling identifies emerging oncogenic pathways operating in extranodal NK/T-cell lymphoma, nasal type. Blood (2010) 115:1226–37. doi: 10.1182/blood-2009-05-221275
16. Iqbal J, Weisenburger DD, Chowdhury A, Tsai MY, Srivastava G, Greiner TC, et al. Natural killer cell lymphoma shares strikingly similar molecular features with a group of non-hepatosplenic gammadelta T-cell lymphoma and is highly sensitive to a novel aurora kinase A inhibitor in vitro. Leukemia (2011) 25:348–58. doi: 10.1038/leu.2010.255
17. Ng SB, Selvarajan V, Huang G, Zhou J, Feldman AL, Law M, et al. Activated oncogenic pathways and therapeutic targets in extranodal nasal-type NK/T cell lymphoma revealed by gene expression profiling. J Pathol. (2011) 223:496–510. doi: 10.1002/path.2823
18. Iqbal J, Kucuk C, Deleeuw RJ, Srivastava G, Tam W, Geng H, et al. Genomic analyses reveal global functional alterations that promote tumor growth and novel tumor suppressor genes in natural killer-cell malignancies. Leukemia (2009) 23:1139–51. doi: 10.1038/leu.2009.3
19. Coppo P, Gouilleux-Gruart V, Huang Y, Bouhlal H, Bouamar H, Bouchet S, et al. STAT3 transcription factor is constitutively activated and is oncogenic in nasal-type NK/T-cell lymphoma. Leukemia (2009) 23:1667–78. doi: 10.1038/leu.2009.91
20. Huang L, Liu D, Wang N, Ling S, Tang Y, Wu J, et al. Integrated genomic analysis identifies deregulated JAK/STAT-MYC-biosynthesis axis in aggressive NK-cell leukemia. Cell Res. (2018) 28:172–86. doi: 10.1038/cr.2017.146
21. Lee S, Park HY, Kang SY, Kim SJ, Hwang J, Lee S, et al. Genetic alterations of JAK/STAT cascade and histone modification in extranodal NK/T-cell lymphoma nasal type. Oncotarget (2015) 6:17764–76. doi: 10.18632/oncotarget.3776
22. Selvarajan V, Osato M, Nah GSS, Yan J, Chung TH, Voon DC, et al. RUNX3 is oncogenic in natural killer/T-cell lymphoma and is transcriptionally regulated by MY. Leukemia (2017) 31:2219–27. doi: 10.1038/leu.2017.40
23. Yan J, Ng SB, Tay JL, Lin B, Koh TL, Tan J, et al. EZH2 overexpression in natural killer/T-cell lymphoma confers growth advantage independently of histone methyltransferase activity. Blood (2013) 121:4512–20. doi: 10.1182/blood-2012-08-450494
24. Yan J, Li B, Lin B, Lee PT, Chung TH, Tan J, et al. EZH2 phosphorylation by JAK3 mediates a switch to noncanonical function in natural killer/T-cell lymphoma. Blood (2016) 128:948–58. doi: 10.1182/blood-2016-01-690701
25. Nairismagi M, Gerritsen ME, Li ZM, Wijaya GC, Chia BKH, Laurensia Y, et al. Oncogenic activation of JAK3-STAT signaling confers clinical sensitivity to PRN371, a novel selective and potent JAK3 inhibitor, in natural killer/T-cell lymphoma. Leukemia (2018) 32:1147–56. doi: 10.1038/s41375-017-0004-x
26. Ng SB, Ohshima K, Selvarajan V, Huang G, Choo SN, Miyoshi H, et al. Epstein-Barr virus-associated T/natural killer-cell lymphoproliferative disorder in children and young adults has similar molecular signature to extranodal nasal natural killer/T-cell lymphoma but shows distinctive stem cell-like phenotype. Leuk Lymphoma (2015) 56:2408–15. doi: 10.3109/10428194.2014.983099
27. Jost PJ, Ruland J. Aberrant NF-κB signaling in lymphoma: mechanisms, consequences, and therapeutic implications. Blood (2007) 109:2700–7. doi: 10.1182/blood-2006-07-025809
28. Liu X, Wang B, Ma X, Guo Y. NF-kappaB activation through the alternative pathway correlates with chemoresistance and poor survival in extranodal NK/T-cell lymphoma, nasal type. Jpn J Clin Oncol. (2009) 39:418–24. doi: 10.1093/jjco/hyp037
29. Dutta-Simmons J, Zhang Y, Gorgun G, Gatt M, Mani M, Hideshima T, et al. Aurora kinase A is a target of Wnt/beta-catenin involved in multiple myeloma disease progression. Blood (2009) 114:2699–708. doi: 10.1182/blood-2008-12-194290
30. Yang H, Ou CC, Feldman RI, Nicosia SV, Kruk PA, Cheng JQ. Aurora-A kinase regulates telomerase activity through c-Myc in human ovarian and breast epithelial cells. Cancer Res. (2004) 64:463–7. doi: 10.1158/0008-5472.CAN-03-2907
31. Murakami M, Hashida Y, Imajoh M, Maeda A, Kamioka M, Senda Y, et al. PCR array analysis of gene expression profiles in chronic active Epstein-Barr virus infection. Microbes Infect. (2014) 16:581–6. doi: 10.1016/j.micinf.2014.04.004
32. Hu X, Ivashkiv LB. Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases. Immunity (2009) 31:539–50. doi: 10.1016/j.immuni.2009.09.002
33. Saha B, Jyothi Prasanna S, Chandrasekar B, Nandi D. Gene modulation and immunoregulatory roles of interferon gamma. Cytokine (2010) 50:1–14. doi: 10.1016/j.cyto.2009.11.021
34. Zhang Y, Ohyashiki JH, Takaku T, Shimizu N, Ohyashiki K. Transcriptional profiling of Epstein-Barr virus (EBV) genes and host cellular genes in nasal NK/T-cell lymphoma and chronic active EBV infection. Br J Cancer (2006) 94:599–608. doi: 10.1038/sj.bjc.6602968
35. Razaghi A, Heimann K, Schaeffer PM, Gibson SB. Negative regulators of cell death pathways in cancer: perspective on biomarkers and targeted therapies. Apoptosis (2018) 23:93–112. doi: 10.1007/s10495-018-1440-4
36. Quintanilla-Martinez L, Kremer M, Keller G, Nathrath M, Gamboa-Dominguez A, Meneses A, et al. p53 Mutations in nasal natural killer/T-cell lymphoma from Mexico: association with large cell morphology and advanced disease. Am J Pathol. (2001) 159:2095–105. doi: 10.1016/S0002-9440(10)63061-1
37. Marechal A, Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol. (2013) 5:a012716. doi: 10.1101/cshperspect.a012716
38. Liu A, Takakuwa T, Luo WJ, Fujita S, Aozasa K. Alterations in ATR in nasal NK/T-cell lymphoma and chronic active Epstein-Barr virus infection. Cancer Sci. (2006) 97:605–10. doi: 10.1111/j.1349-7006.2006.00226.x
39. Boussiotis VA. Molecular and biochemical aspects of the PD-1 checkpoint pathway. N Engl J Med. (2016) 375:1767–78. doi: 10.1056/NEJMra1514296
40. Hu B, Oki Y. Novel immunotherapy options for extranodal NK/T-Cell Lymphoma. Front Oncol. (2018) 8:139. doi: 10.3389/fonc.2018.00139
41. Bi XW, Wang H, Zhang WW, Wang JH, Liu WJ, Xia ZJ, et al. PD-L1 is upregulated by EBV-driven LMP1 through NF-kappaB pathway and correlates with poor prognosis in natural killer/T-cell lymphoma. J Hematol Oncol. (2016) 9:109. doi: 10.1186/s13045-016-0341-7
42. Lim ST, Song T, Lim JQ, Laurensia Y, Pang JWL, Nagarajan S, et al. Oncogenic activation of STAT3 pathway drives PD-L1 expression in natural killer/T cell lymphoma. J Clin Oncol. (2017) 35:7549. doi: 10.1200/JCO.2017.35.15_suppl.7549
43. Wang L, Wang H, Li PF, Lu Y, Xia ZJ, Huang HQ, et al. CD38 expression predicts poor prognosis and might be a potential therapy target in extranodal NK/T cell lymphoma, nasal type. Ann Hematol. (2015) 94:1381–8. doi: 10.1007/s00277-015-2359-2
44. Mustafa N, Nee HFA, Lee XTJ, Jin W, Yu Y, Chen Y, et al. Daratumumab efficiently targets NK/T cell lymphoma with high CD38 expression. Blood (2017) 130:2814.
45. Burr ML, Sparbier CE, Chan YC, Williamson JC, Woods K, Beavis PA, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature (2017) 549:101–5. doi: 10.1038/nature23643
46. Zhong H, Hu X, Janowski AB, Storch GA, Su L, Cao L, et al. Whole transcriptome profiling reveals major cell types in the cellular immune response against acute and chronic active Epstein-Barr virus infection. Sci Rep. (2017) 7:17775. doi: 10.1038/s41598-017-18195-z
47. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
48. Hicklin J, Ellis LM. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J Clin Oncol. (2005) 23:1011–27. doi: 10.1200/JCO.2005.06.081
49. Jørgensen JM, Sørensen FB, Bendix K, Nielsen JL, Funder A, Karkkainen MJ, et al. Expression level, tissue distribution pattern, and prognostic impact of vascular endothelial growth factors VEGF and VEGF-C and their receptors Flt-1, KDR, and Flt-4 in different subtypes of non-Hodgkin lymphomas. Leuk Lymphoma (2009) 50:1647–60. doi: 10.1080/10428190903156729
50. Ito Y, Shibata-Watanabe Y, Ushijima Y, Kawada J, Nishiyama Y, Kojima S, et al. Oligonucleotide microarray analysis of gene expression profiles followed by real-time reverse-transcriptase polymerase chain reaction assay in chronic active Epstein-Barr virus infection. J Infect Dis. (2008) 197:663–6. doi: 10.1086/527330
51. Ohga S, Nomura A, Takada H, Tanaka T, Furuno K, Takahata Y, et al. Dominant expression of interleukin-10 and transforming growth factor-beta genes in activated T-cells of chronic active Epstein-Barr virus infection. J Med Virol. (2004) 74:449–58. doi: 10.1002/jmv.20197
52. Ito T, Kawazu H, Murata T, Iwata S, Arakawa S, Sato Y, et al. Role of latent membrane protein 1 in chronic active Epstein-Barr virus infection-derived T/NK-cell proliferation. Cancer Med. (2014) 3:787–95. doi: 10.1002/cam4.256
53. Westin JR. Status of PI3K/Akt/mTOR pathway inhibitors in lymphoma. Clin Lymphoma Myeloma Leuk. (2014) 14:335–42. doi: 10.1016/j.clml.2014.01.007
54. Aster JC, Blacklow SC, Pear WS. Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J Pathol. (2011) 223:262–73. doi: 10.1002/path.2789
55. Beaulieu M, Bezman NA, Lee JE, Matloubian M, Sun JC, Lanier LL. MicroRNA function in NK-cell biology. Immunol Rev. (2013) 253:40–52. doi: 10.1111/imr.12045
56. Fujiwara S, Kimura H, Imadome K, Arai A, Kodama E, Morio T, et al. Current research on chronic active Epstein-Barr virus infection in Japan. Pediatr Int. (2014) 56:159–66. doi: 10.1111/ped.12314
57. Ng SB, Yan J, Huang G, Selvarajan V, Tay JL, Lin B, et al. Dysregulated microRNAs affect pathways and targets of biologic relevance in nasal-type natural killer/T-cell lymphoma. Blood (2011) 118:4919–29. doi: 10.1182/blood-2011-07-364224
58. Motsch N, Alles J, Imig J, Zhu J, Barth S, Reineke T, et al. MicroRNA profiling of Epstein-Barr virus-associated NK/T-cell lymphomas by deep sequencing. PLoS ONE (2012) 7:e42193. doi: 10.1371/journal.pone.0042193
59. Yamanaka Y, Tagawa H, Takahashi N, Watanabe A, Guo YM, Iwamoto K, et al. Aberrant overexpression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer-cell lymphoma/leukemia. Blood (2009) 114:3265–75. doi: 10.1182/blood-2009-06-222794
60. Paik JH, Jang JY, Jeon YK, Kim WY, Kim TM, Heo DS, et al. MicroRNA-146a downregulates NFkappaB activity via targeting TRAF6 and functions as a tumor suppressor having strong prognostic implications in NK/T cell lymphoma. Clin Cancer Res. (2011) 17:4761–71. doi: 10.1158/1078-0432.CCR-11-0494
61. Guo HQ, Huang GL, Guo CC, Pu XX, Lin TY. Diagnostic and prognostic value of circulating miR-221 for extranodal natural killer/T-cell lymphoma. Dis Markers (2010) 29:251–8. doi: 10.1155/2010/474692
62. Kawano Y, Iwata S, Kawada J, Gotoh K, Suzuki M, Torii Y, et al. Plasma viral microRNA profiles reveal potential biomarkers for chronic active Epstein-Barr virus infection. J Infect Dis. (2013) 208:771–9. doi: 10.1093/infdis/jit222
63. Sato E, Ohga S, Kuroda H, Yoshiba F, Nishimura M, Nagasawa M, et al. Allogeneic hematopoietic stem cell transplantation for Epstein-Barr virus-associated T/natural killer-cell lymphoproliferative disease in Japan. Am J Hematol. (2008) 83:721–7. doi: 10.1002/ajh.21247
64. Kawa K, Sawada A, Sato M, Okamura T, Sakata N, Kondo O, et al. Excellent outcome of allogeneic hematopoietic SCT with reduced-intensity conditioning for the treatment of chronic active EBV infection. Bone Marrow Transplant. (2011) 46:77–83. doi: 10.1038/bmt.2010.122
65. Kimura H, Ito Y, Kawabe S, Gotoh K, Takahashi Y, Kojima S, et al. EBV-associated T/NK-cell lymphoproliferative diseases in nonimmunocompromised hosts: prospective analysis of 108 cases. Blood (2012) 119:673–86. doi: 10.1182/blood-2011-10-381921
66. Quintanilla-Martinez L, Ridaura C, Nagl F, Saez-de-Ocariz M, Duran-McKinster C, Ruiz-Maldonado R, et al. Hydroa vacciniforme-like lymphoma: a chronic EBV+lymphoproliferative disorder with risk to develop a systemic lymphoma. Blood (2013) 122:3101–10. doi: 10.1182/blood-2013-05-502203
67. Ng SB, Ohshima K, Selvarajan V, Huang G, Choo SN, Miyoshi H, et al. Prognostic implication of morphology, cyclinE2, and proliferation in EBV-associated T/NK lymphoproliferative disease in non-immunocompromised hosts. Orphanet J Rare Dis. (2014) 9:165. doi: 10.1186/s13023-014-0165-x
68. Kim SJ, Choi JY, Hyun SH, Ki CS, Oh D, Ahn YC, et al. Risk stratification on the basis of Deauville score on PET-CT and the presence of Epstein-Barr virus DNA after completion of primary treatment for extranodal natural killer/T-cell lymphoma, nasal type: a multicentre, retrospective analysis. Lancet Haematol. (2015) 2:e66–74. doi: 10.1016/S2352-3026(15)00002-2
69. Kwong YL, Chan TSY, Tan D, Kim SJ, Poon LM, Mow B, et al. PD1 blockade with pembrolizumab is highly effective in relapsed or refractory NK/T-cell lymphoma failing l-asparaginase. Blood (2017) 129:2437–42. doi: 10.1182/blood-2016-12-756841
70. Lokhorst HM, Plesner T, Laubach JP, Nahi H, Gimsing P, Hansson M, et al. Targeting CD38 with daratumumab monotherapy in multiple myeloma. N Engl J Med. (2015) 373:1207–19. doi: 10.1056/NEJMoa1506348
71. Hari P, Raj RV, Olteanu H. Targeting CD38 in refractory extranodal natural killer cell-T-cell lymphoma. N Engl J Med. (2016) 375:1501–2. doi: 10.1056/NEJMc1605684
72. Sim SH, Kim S, Kim TM, Jeon YK, Nam SJ, Ahn YO, et al. Novel JAK3-activating mutations in extranodal NK/T-cell lymphoma, nasal type. Am J Pathol. (2017) 187:980–6. doi: 10.1016/j.ajpath.2017.01.004
73. Zhang Y, Xu W, Liu H, Li J. Therapeutic options in peripheral T cell lymphoma. J Hematol Oncol. (2016) 9:37. doi: 10.1186/s13045-016-0267-0
74. Tang T, Tay K, Tao M, Quek RHH, Farid M, Lim ST. A phase II study of Bortezomib-GIFOX (Gemcitabine, Ifosfamide, Oxaliplatin) in patients with newly diagnosed natural-killer/T-cell lymphoma. Blood (2016) 128:5353.
Keywords: Epstein–Barr virus, extranodal NK/T cell lymphoma, chronic active EBV infection, T cell lymphoma, RNA
Citation: de Mel S, Tan JZ-C, Jeyasekharan AD, Chng W-J and Ng S-B (2019) Transcriptomic Abnormalities in Epstein Barr Virus Associated T/NK Lymphoproliferative Disorders. Front. Pediatr. 6:405. doi: 10.3389/fped.2018.00405
Received: 12 September 2018; Accepted: 06 December 2018;
Published: 17 January 2019.
Edited by:
Shigeyoshi Fujiwara, National Center for Child Health and Development (NCCHD), JapanReviewed by:
Takayuki Murata, Fujita Health University, JapanLisa Renee Forbes, Baylor College of Medicine, United States
Copyright © 2019 de Mel, Tan, Jeyasekharan, Chng and Ng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siok-Bian Ng, patnsb@nus.edu.sg
 Sanjay de Mel
Sanjay de Mel Joshua Zhi-Chien Tan2
Joshua Zhi-Chien Tan2  Siok-Bian Ng
Siok-Bian Ng