- 1Department of General and Visceral Surgery, Surgery Center, Ulm University Hospital, Ulm, Germany
- 2Institute for Pharmaceutical Chemistry, Christian Albrechts University, Kiel, Germany
- 3Department of Organic Synthesis, Ural Federal University, Ekaterinburg, Russia
Members of the highly conserved and ubiquitously expressed pleiotropic CK1 family play major regulatory roles in many cellular processes including DNA-processing and repair, proliferation, cytoskeleton dynamics, vesicular trafficking, apoptosis, and cell differentiation. As a consequence of cellular stress conditions, interaction of CK1 with the mitotic spindle is manifold increased pointing to regulatory functions at the mitotic checkpoint. Furthermore, CK1 is able to alter the activity of key proteins in signal transduction and signal integration molecules. In line with this notion, CK1 is tightly connected to the regulation and degradation of β-catenin, p53, and MDM2. Considering the importance of CK1 for accurate cell division and regulation of tumor suppressor functions, it is not surprising that mutations and alterations in the expression and/or activity of CK1 isoforms are often detected in various tumor entities including cancer of the kidney, choriocarcinomas, breast carcinomas, oral cancer, adenocarcinomas of the pancreas, and ovarian cancer. Therefore, scientific effort has enormously increased (i) to understand the regulation of CK1 and its involvement in tumorigenesis- and tumor progression-related signal transduction pathways and (ii) to develop CK1-specific inhibitors for the use in personalized therapy concepts. In this review, we summarize the current knowledge regarding CK1 regulation, function, and interaction with cellular proteins playing central roles in cellular stress-responses and carcinogenesis.
The CK1 Family
Members of the CK1 (formerly named casein kinase 1) family were among the first kinases described in literature (1). Although the milk protein component casein is not a physiological substrate for CK1, it reflects its preference for serine or threonine residues N-terminally flanked by already phosphorylated amino acid residues or acidic amino acids (2–7). Seven distinct genes encoding mammalian CK1 isoforms α, β, γ1, γ2, γ3, δ, and ε as well as various post-transcriptionally processed splice variants (transcription variants; TV) have been characterized (except for β all are expressed in humans). The closest relatives to the CK1 family are tau tubulin kinases 1 and 2 (TTBK1/2) and the vaccinia-related kinases 1–3 (VRK1-3) (Figure 1A). All CK1 isoforms are highly conserved within their kinase domains (51–98% identical) while the highly related isoforms CK1δ and ε display the highest homology. However, CK1 family members differ significantly in length and primary structure of their regulatory non-catalytic C-terminal domains, resulting in molecular weights ranging from 32 kDa (CK1α) to 52.2 kDa (CK1γ3) (Figure 1B) (5, 8–16). Meanwhile, CK1 homologous proteins have also been isolated from yeast, basidiomycetes, plants, algae, and protozoa (9, 15, 17–23). Since recognition motifs for CK1 are found on most cellular proteins, more than 140 in vitro and in vivo substrates have been reported thus far (see CK1 Substrate Specificity and Table 1). Therefore, in a cellular context a tight regulation of CK1 activity and expression is indispensable. Known general mechanisms for CK1 regulation include (i) phosphorylation by inhibitory autophosphorylation and/or (ii) phosphorylation by other cellular protein kinases, and (iii) interaction with cellular proteins or subcellular sequestration (see Regulation of CK1 Activity). Based on the broad spectrum of target proteins, CK1 family members are involved in modulating a variety of cellular functions: in immune response and inflammation (see CK1 in Immune Response and Inflammation), in spindle and centrosome-associated processes (see Interaction of CK1 with Centrosomes, Tubulin, and Microtubule-Associated Proteins), in DNA damage-related signal transduction (see CK1 in DNA Damage-Related Signal Transduction), in circadian rhythm (see CK1 in Circadian Rhythm and its Connections to Stress Response), and in apoptosis (see CK1-Signaling in Apoptotic Pathways). Consequently, a deregulation or dysfunction of CK1 in pathways responsible for regulation of growth, proliferation, and apoptosis may result in pathological conditions (see CK1 and the Wnt Pathway, CK1 in the Hedgehog Pathway to CK1 in the Hippo Pathway), such as tumorigenesis (see CK1-Related Tumorigenic Functions and CK1 in Metastatic Processes) or neurological diseases. Therefore, interest in CK1 isoforms as new drug targets has enormously increased within the last 15 years and led to development of several CK1-specific inhibitors (see CK1-Specific Inhibitors).
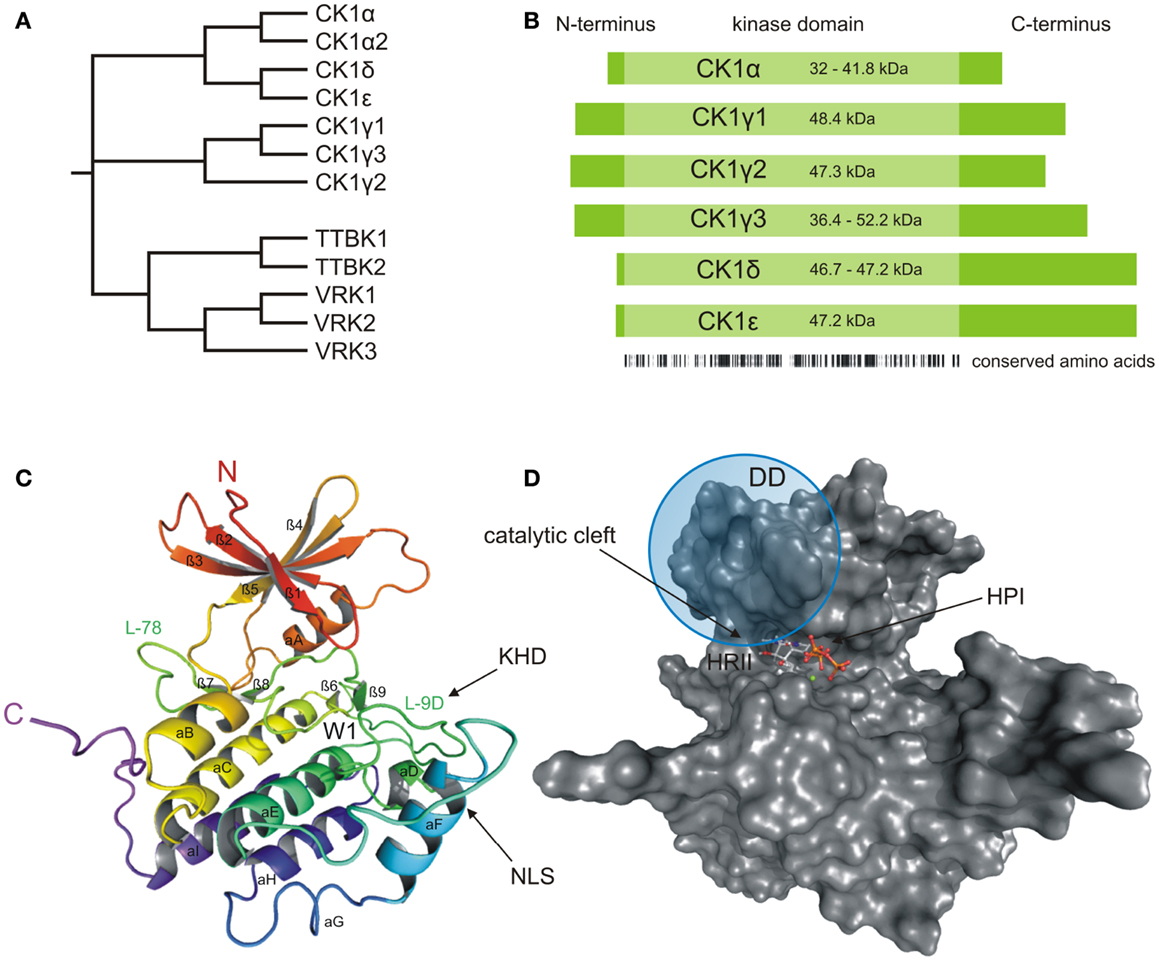
Figure 1. Structural presentation of CK1δ. (A) Phylogenetic relation between CK1 isoforms of Homo sapiens (CK1α, γ1–3, δ, and ε) and other members of the human CK1 family (TTBK1–2, VRK1–3). (B) Schematic alignment of human CK1 isoforms α, γ1–3, δ, and ε. Their molecular weight varies between 32 (CK1α) and 52.2 kDa (CK1γ3). In case transcription variants have been reported for one isoform, the molecular weight is given as range from the smallest to the largest variant. All CK1 isoforms are highly conserved within their kinase domains (light green box, 286 aa), but differ within their variable N- (4–40 aa) and C-terminal (39–122 aa) non-catalytic domains (dark green boxes) [according to Knippschild et al. (333)]. Ribbon (C) and surface (D) diagram of the molecular structure of CK1δ (PDB code 4HGT) modeled in complex with Mg2+-ATP at a resolution of 1.80 Å. The nomenclature is adapted from Xu et al. (24) and Longenecker et al. (25). Until today, crystal structures of human CK1 isoforms γ1 (PDB code 2CMW), γ2 (2C47), γ3 (2CHL, 2IZR, 2IZS, 2IZT, 2IZU, 4HGL, 4HGS, 4G16, 4G17), δ (4KB8, 4KBA, 4KBC, 4KBK, 4HNF, 3UYS, 3UYT, 3UZP), and ε (4HNI, 4HOK) are accessible as well. For reasons of clarity, we focused on CK1δ exemplarily, due to its superior relevance. The catalytic domain folds into two lobes primarily containing strands (N-terminal), respectively helices (C-terminal) forming a catalytic cleft between that represents the ATP binding pocket as well as a substrate binding site. KHD indicates the kinesin homology domain within L-9D. DD refers to a putative dimerization domain containing various amino acids of β1, β2, β5, L-5B, β7, and αB, whereas NLS displays a putative nuclear localization signal sequence at the junction between L-EF and αF. A tungstate molecule binding site identifies a specific phosphate moiety binding motif (W1). The active site contains a deep hydrophobic pocket (HPI) and a spacious hydrophobic region (HRII) (25–28). All modeling and docking studies were performed using Schrödinger software (Maestro, version 9.3, Schrödinger, LLC, New York, NY, 2012; Glide, version 5.8, Schrödinger, LLC, New York, NY, 2012). The illustration of modeling results was generated by the PyMOL Molecular Graphics System (Version 1.5.0.4, LLC) (29).

Table 1. Reported substrates for CK1 family members and reported in vitro and in vivo substrates of CK1 family members of several species.
CK1 Structure and Domains
As a member of the superfamily of serine/threonine-specific kinases, CK1 represents the typical bi-lobal structure, which includes a smaller N-terminal lobe, primarily consisting of β-sheets, and a larger, mainly α-helical C-terminal lobe. The two lobes are connected by a hinge region forming a catalytic cleft for substrate and ATP binding (Figures 1C,D) (24, 25). In comparison to the general structural features of protein kinases, a prominent α-helix (αA-helix) within the N-terminal region is crucial for conformational regulation of kinase activity. A conserved glycine-rich loop (P-loop, bridging strands β1 and β2) forms the ceiling of the ATP active site and contributes to coordination of the γ-phosphate moiety of ATP (30). Contributing to structure-based inhibitor design, another loop (L-78) in close proximity to the hinge region has been demonstrated to trigger CK1 inhibitor selectivity (31). Within the C-terminal region, a specific phosphate moiety binding motif (W1) has been identified affording recognition of phosphorylated protein substrates and is further believed to be involved in CK1 regulatory interactions (9, 24, 25). In addition, a kinesin homology domain (KHD) within the T-loop (L-9D) and a putative dimerization domain (DD, containing various amino acids of strands β1, β2, β5, hinge region, β7, and αB) can be found inside the catalytic domain of CK1δ (Figures 1C and 2) (26, 32–34). The KHD is thought to support the interaction of CK1 isoforms with components of the cytoskeleton as this domain has been shown to be necessary for the interaction of kinesins with microtubules (MT) (26, 32–34). Furthermore, a putative nuclear localization signal sequence (NLS) at the junction between L-EF and αF has been reported to affect substrate binding (Figure 1C). The present NLS however seems to be not sufficient for nuclear localization of CK1δ because only CK1αL variants, carrying an additional NLS in the L-exon, are able to localize to the nucleus (35).The L-9D loop represents the homolog of the so-called activation-loop identified in other protein kinases and may therefore play a role in CK1 regulation. Moreover, loops L-9D and L-EF may be of importance in substrate recognition (Figure 1C) (24–27). The ATP active site itself mainly consists of a deep hydrophobic pocket (HPI, selectivity pocket) lined by the gatekeeper (Met-82 in CK1δ) and a second spacious hydrophobic region (HRII) adjacent to the hinge region as well as sugar and phosphate binding domains (Figure 1D) (31).
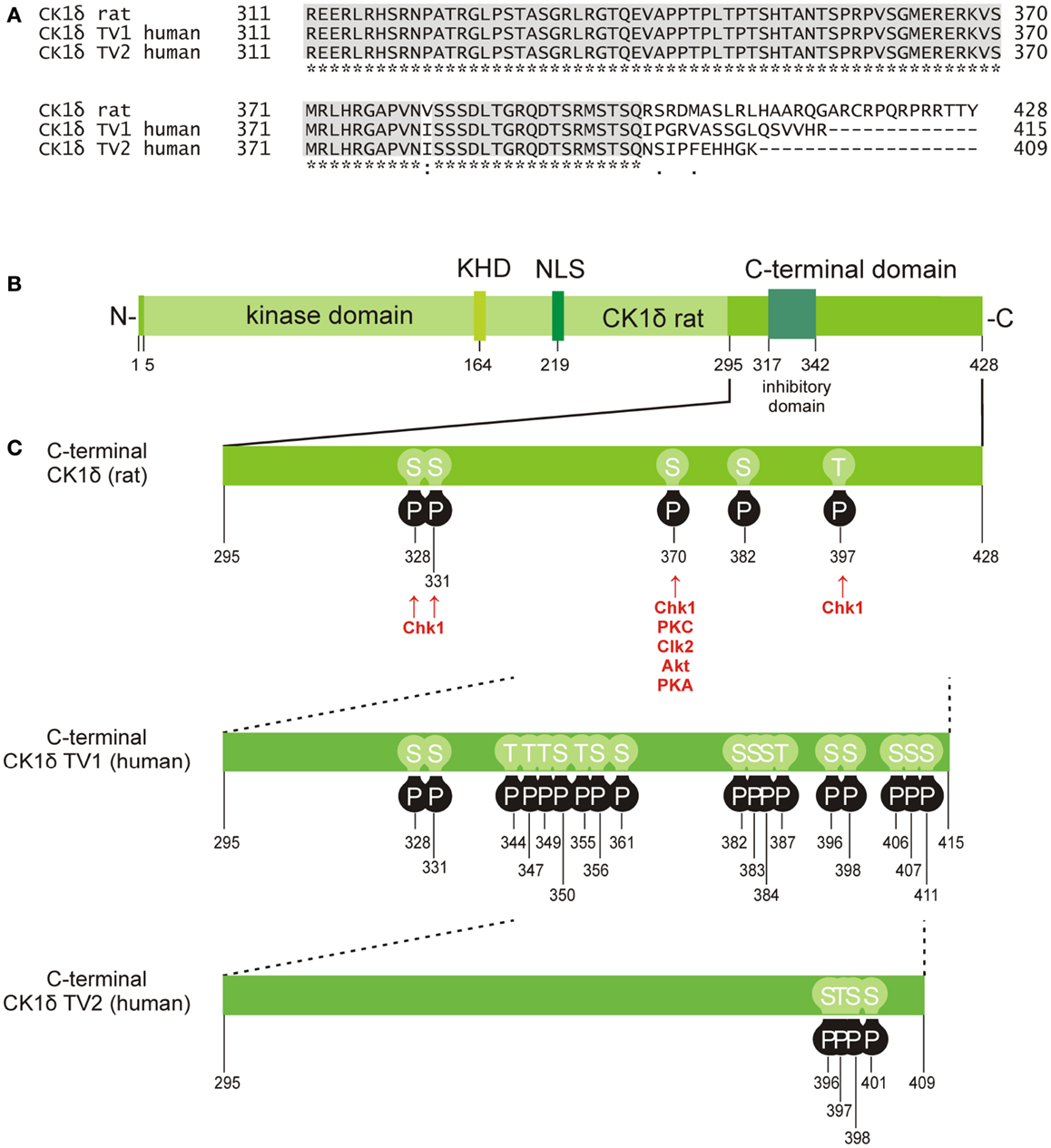
Figure 2. Phosphorylation sites located in the C-terminal domain of CK1δ. (A) Alignment of the rat CK1δ C-terminal sequence with the human CK1δ transcription variants 1 and 2 C-terminal sequences (accession numbers L07578, NM001893, and NM139062, respectively) generated by using the program ClustalW (36, 37), showing conserved amino acids (gray) and obvious differences in the C-terminal domain beyond amino acid 399. (B) Domain structure of rat CK1δ (NLS: nuclear localization signal, KHD: kinesin homology domain). (C) Phosphorylation sites in the C-terminal regulatory domains of CK1δ rat and human transcription variants 1 and 2, that have so far been confirmed experimentally (38–53). Kinases identified for phosphorylation of the C-terminal domain are shown for rat CK1δ (38, 39).
CK1 Substrate Specificity
Belonging to the group of acidotropic protein kinases, CK1 family members mainly recognize substrates containing acidic or phosphorylated amino acid residues. The canonical consensus sequence for CK1 protein kinases is represented by the motif pSer/Thr-X-X-(X)-Ser/Thr whereas pSer/Thr indicates a phosphorylated serine or threonine residue. However, CK1 not only relies on phospho-primed motifs since the phospho-serine or phospho-threonine can also be replaced by an agglomeration of negatively charged acidic amino acids (2–7). In addition, non-canonical consensus sequences for CK1 family members have been described such as the SLS motif, found in β-catenin and nuclear factor of activated T-cells (NFAT), or the motif Lys/Arg-X-Lys/Arg-X-X-Ser/Thr occurring in sulfatide and cholesterol-3-sulfate (SCS) binding proteins (54, 55). Generally, substrate recognition motifs for CK1 protein kinases are massively distributed on cellular proteins. At present, more than 140 in vitro and in vivo substrates for CK1 isoforms have been reported, underlining its pleiotropic character (Table 1).
Regulation of CK1 Activity
Although members of the CK1 family are ubiquitously expressed, their expression levels differ depending on tissue and cell type (34, 183, 184). Certain factors seem to change the expression and activity of CK1, such as stimulation with insulin (185) or gastrin (140), viral transformation (186), treatment with topoisomerase inhibitors or other small molecules like calotropin (187), γ-irradiation (188), or altered membrane concentrations of phosphatidylinositol-4,5-bisphosphate (PIP2) (172). At the protein level, certain mechanisms regulating CK1 activity have been identified: structure-related regulation, subcellular localization, interaction with other proteins, and post-translational modifications.
In X-ray crystallography, CK1δ was found to form dimers. In the dimeric form, the adenine binding domain is occupied by the specific intramolecular contacts of the dimerization domain. As a consequence, ATP is excluded from the active center of the kinase. Therefore, formation of homodimers could possibly have a negative regulatory effect on CK1δ kinase activity in vivo (26). This hypothesis is supported by further observations: the expression of a mutant CK1δ with impaired kinase activity lead to down-regulation of endogenous CK1δ activity in a dominant-negative way in simian virus 40 (SV40)-transformed cell lines as well as to changes in mammary tumorigenesis in WAP-mutCK1δ/WAP-T bi-transgenic mice (189, 190).
Appropriate sequestration of CK1 proteins to particular cellular compartments is crucial for access to their pool of substrates (21, 191, 192). As an example, in Saccharomyces cerevisiae kinase activity of C-terminal deletion mutants of membrane-bound YCK1 and YCK2 could only be rescued by replacing the nuclear localization signal of the CK1 homolog Hrr25 with a prenylation motif, which is required for plasma membrane localization. Conversely, loss of Hrr25 function after deletion of its NLS could only be rescued by replacing the prenylation motif in YCK1 and YCK2 with a NLS. These observations led to the conclusion that merely partial cellular overlap of these three isoforms is not enough to rescue the deletion phenotype (192). In experiments using a CK1δ kinase-dead mutant, it has been shown that not only the existence of the kinase domain, but also the catalytic activity of the protein is essential for its appropriate subcellular localization (193). Additionally, a study designed to identify binding partners, which recruit CK1 to Alzheimer’s disease (AD) ubiquitinated lesions identified a dysbindin structural homolog that interacts with CK1γ, δ, and ε, and in the case of CK1δ it has been shown to be a concentration-dependent inhibitor (194).
It is very common to find certain motifs in proteins that act as scaffolds, which direct the proper positioning of protein complexes. It has also been suggested that such scaffolds additionally exert complex allosteric control of their partners thereby regulating their activity [reviewed in Cheong and Virshup (195) and Good et al. (196)]. In general, proteins that function as scaffolds tether members of signaling pathways into complexes thereby increasing the interaction efficiency between partner molecules (196, 197). In the case of CK1, these scaffolds have an important regulatory role because they might change the affinity of CK1 isoforms for their substrates as well as the rate of phosphorylation and activation of CK1 kinase activity over the basal level (196, 198). In fact, protein scaffolds have been already found to exert substantial control over different kinase-mediated signaling pathways [reviewed in Brown et al. (199)], though they are not limited to the coordination of kinase cascades (196). Examples for such protein scaffolds include the centrosomal and Golgi N-kinase anchoring protein (CG-NAP), also known as A-kinase anchoring protein 450 (AKAP450) (191) and the DEAD-box RNA helicase DDX3, which has been previously identified as scaffolding adaptor that directly activates the kinase IκB (200). AKAP450 specifically interacts with CK1δ and ε and recruits them to the centrosome, where they can exert centrosome-specific functions coupled to the cell cycle. This interaction is confirmed by the ability of AKAP450 to re-localize CK1δ at the plasma membrane, when it itself is attached to the membrane (191). Recently, it has been suggested that the interaction of CK1δ with AKAP450 is necessary to mediate primary ciliogenesis (201). In addition, evidence is increasing that in Wnt-signaling CK1 activity depends on DDX3 as a co-factor. DDX3 directly interacts with CK1ε in a Wnt-dependent manner, and promotes phosphorylation of Disheveled (DVL) (103). DDX3 can therefore be seen as regulatory subunit of CK1 isoforms with the potential to increase the activity of CK1α, γ2, δ, and ε by up to five orders of magnitude (103). Since CK1 isoforms have been shown to phosphorylate DDX3, it could be speculated that CK1 isoforms might also play a role in regulating the functions of DDX3 (103).
Finally, CK1 activity can furthermore be regulated by post-translational modifications, mainly represented by reversible phosphorylation either through autophosphorylation or site-specific phosphorylation mediated by cellular kinases. Within the regulatory C-terminal domains of CK1δ and ε, sequences with the motif pSer/Thr-X-X-Y (Y: any amino acid except serine or threonine) can be generated by autophosphorylation events and can consecutively act as pseudo-substrates blocking the catalytic center of the kinase (202–205). By using CK1δ truncation mutants, Ser-318, Thr-323, Ser-328, Thr-329, Ser-331, and Thr-337 were detected as candidate sites for intramolecular autophosphorylation. Although not all of them influenced kinase activity, truncation of the C-terminal part up to amino acid (aa) 317 significantly enhanced activity of CK1δ (204). For CK1ε amino acid residues Ser-323, Thr-325, Thr-334, Thr-337, Ser-368, Ser-405, Thr-407, and Ser-408 within the C-terminal domain are considered to be potential autophosphorylation sites (203). C-terminal inhibitory autophosphorylation could also be demonstrated for CK1γ1-3 as well as for CK1α and its splice variants CK1αL and CK1αS (16, 206).
Apart from intramolecular autophosphorylation, CK1 isoforms are also phosphorylated by other kinases. In the case of CK1δ, phosphorylation by PKA (cAMP-dependent protein kinase), Akt (protein kinase B), CLK2 (CDC-like kinase 2), protein kinase C isoform α (PKCα), and Chk1 (checkpoint kinase 1) has been demonstrated (38, 39) (Figure 2). PKA could be further characterized as a major CK1δ C-terminal targeting kinase predominantly phosphorylating Ser-370 both in vitro and in vivo. Mutation of Ser-370 to alanine increased kinase activity in vitro and enhanced formation of an ectopic dorsal axis during embryonic development of Xenopus laevis (39). More recently, Chk1 has been demonstrated to phosphorylate CK1δ at serine residues 328, 331, and 370, as well as threonine residue 397. Mutations at these sites proved to significantly increase kinase activity (38). Moreover, several residues in the C-terminal domain of CK1δ were found in a phosphorylated state in large-scale mass spectrometry analyses. However, the kinases responsible for the detected phosphorylation events were not specified (Figure 2 and references therein).
Generally, dephosphorylation of CK1 by serine/threonine-specific protein phosphatases or low levels of H2O2 result in an increase of kinase activity (202, 203, 207). Proteolytic cleavage of the C-terminal domain also results in multiple increase of CK1 kinase activity in vitro (28, 202, 204). In addition, neddylation of CK1α seems to be involved in CK1 regulation (208).
CK1 in Stress-Related Cellular Functions
In response to stress situations like mechanical damage, toxin exposure, or environmental stress exposure, cells experience a variety of molecular changes, which are generally referred to as cellular stress response. The purpose of these changes is to protect the cell against conditions, which may cause acute damage, but also to build some kind of resistance toward long term unfavorable conditions. In response to extreme temperature or toxic substances, expression of heat shock proteins (Hsp) is transcriptionally increased. Most of these proteins belong to a group of proteins, which are involved in the (un-)folding of other proteins (209). A quite recent report links phosphorylation events mediated by CK1, CK2, and GSK3β to the regulation of Hsp70 and Hsp90. In more detail, phosphorylation of Hsp70 and Hsp90, mediated by these kinases, plays an important role in regulating their binding to co-chaperones like HOP (protein folding activity) and CHIP (ubiquitin ligase activity). In highly proliferative cells, phosphorylated Hsp70 and 90 form complexes with HOP whereas CHIP-binding is prevented by phosphorylation of Hsp70 and 90. Therefore, CK1, CK2, and GSK3β together with the action of phosphatases might be involved in complex regulation of the C-terminal phosphorylation of Hsp70 and Hsp90 and their binding to co-chaperones (78). Moreover, apart from environmental or external stress conditions, cells may also be challenged by stress originating from pathological conditions as in the case of inflammatory or proliferative diseases. A detailed presentation of CK1 isoforms in regulating cellular stress response can be found in the following chapters.
CK1 in Immune Response and Inflammation
By analyzing lymphatic tissues of BALB/c mice, remarkable immunoreactivity of CK1δ and ε in granulocytic and megakaryotic cells as well as in a subpopulation of lymphocytes has been detected (183, 184, 210). Mitogenic activation of T-lymphocytes was accompanied by a significant increase in both CK1δ protein levels and kinase activity (210).
So far, several mechanisms have been reported by which CK1 isoforms might be involved in regulating lymphocyte activation and granulocyte physiology. Transcriptional activators of the NFAT family of proteins play a major role in T-cell activation. Their translocation to the nucleus can be blocked by phosphorylation of numerous sites present in the NFAT regulatory domain (211). Some of these are phosphorylated by various CK1 isoforms (rat liver CK1 and Danio rerio CK1α) with high efficiency. In a two-phase phosphorylation mechanism, first phosphorylation of the non-canonical site Ser-177 is initiated by CK1 binding to a cluster of acidic residues within the sequence of aa 173–218. This event enhances the subsequent phosphorylation of downstream residues in a hierarchical manner (212). In contrast, Okamura and colleagues reported NFAT1 to be phosphorylated by CK1 within the serine-rich region SRR-1 (aa 149–183) after binding of CK1 to a N-terminal motif between aa 1–98 (213).
Upon T-cell receptor engagement dynamic association of CK1α to the CBM (CARMA/BCL10/MALT1) complex has been shown. This complex acts as an NFκB (nuclear factor “kappa-light-chain-enhancer” of activated B cells) activating platform containing the scaffold protein CARMA1, the adaptor protein BCL10, and the paracaspase MALT1. Here, CK1α complex association is linked to NFκB activation, increased cytokine production, and lymphocyte proliferation. However, CK1α was found to be a bi-functional regulator of NFκB signaling since phosphorylation and subsequent inactivation of CARMA1 leads to termination of receptor-induced NFκB activation (159). Just recently, CK1γ1 has been demonstrated to be a negative regulator in innate immunity by directly phosphorylating the NFκB subunit p65 following RIG-I pathway stimulation after RNA virus infection. This phosphorylation event is sufficient to target p65 for its degradation (129). Following immune receptor engagement a signal transduction platform is assembled around the T-cell receptor. This specialized cell–cell junction is known as the immunological synapse whose formation also leads to remodeling of the actin cytoskeleton and to repositioning of the centrosome to the immunological synapse (214). Herein, the polarization process is supported by CK1δ phosphorylating the microtubule plus-end-binding protein 1 (EB1). Formation of CK1δ–EB1 complexes is associated with increased speed of microtubule growth and most likely also with subsequent centrosome translocation in activated T-cells (74).
In granulocytes as well as in solid tumors cell survival is significantly promoted by the transcriptional activator HIF-1 (hypoxia-inducible factor-1), which is able to respond to changes in cellular oxygen levels. HIF-1 is continuously produced and marked for degradation by a hydroxylation step involving oxygen-dependent hydroxylases. Under hypoxic conditions, the continuous destruction of HIF-1 is blocked (215). Additionally, HIF-1 expression and activity can be regulated by oxygen-independent mechanisms resulting in phosphorylation of critical residues in HIF-1 regulatory domains. CK1δ has been identified as one of these kinases able to phosphorylate Ser-247 in the PAS-B (Per-ARNT-Sim-B) domain of HIF-1α. This modification has no effect on HIF-1α stability but affects the formation of the transcriptionally active HIF-1α-ARNT heterodimer, which is seen as an obligatory step prior to DNA binding (216). Therefore, active CK1δ can be seen as negative regulator of HIF-1-mediated cell survival.
Additionally, for the highly CK1δ homologous isoform CK1ε, a major role for transcriptional regulation in granulocytes has been suggested. Along with human granulocytic differentiation, a down-regulation of CK1ε has been observed. Here, active CK1ε was shown to interact with and to stabilize SOCS3 (suppressor of cytokine signaling 3) leading to attenuation of STAT3. Consequently, overexpression of CK1ε inhibited granulocyte-colony-stimulating factor (G-CSF) induced differentiation of myeloid progenitor cells (217).
Interaction of CK1 with Centrosomes, Tubulin, and Microtubule-Associated Proteins
Members of the CK1 family represent central components in the regulation of several cellular functions linked to cell cycle progression, spindle-dynamics, and chromosome segregation. CK1α has been shown to be located at the centrosome, microtubule asters, and the kinetochore (218–220). In addition, CK1δ especially associates with the spindle apparatus during mitosis and directly modulates MT by phosphorylation of α-, β-, and γ-tubulin, thereby exerting stress-induced functions at the spindle apparatus and the centrosome (221, 222). Recently, knockdown of CK1δ by siRNA was reported to inhibit microtubule nucleation at the Golgi apparatus (201). Furthermore, homologs of CK1, such as casein kinase 1-like 6 (CKL6), associate with cortical MT in vivo and phosphorylate tubulin in vitro (223).
In addition to the direct interaction of CK1 with MT, their polymerization and stability can also be regulated by CK1-mediated phosphorylation of microtubule-associated proteins (MAPs) (224). CK1δ regulates microtubule- and spindle-dynamics in response to genotoxic stress in order to maintain genomic stability by site-specific phosphorylation of tubulin, stathmin, and the MAPs MAP4, MAP1A, and tau (32, 63, 219, 225–227) as well as the phosphorylation of Sid4 that delays cytokinesis (75). An abnormal hyperphosphorylation of tau by CK1δ can lead to microtubule destabilization and is associated with the pathogenesis of AD (220, 225, 227).
Recent studies provide evidence that CK1 influences dynein-dependent transport along MT. For instance, CK1ε phosphorylates dynein intermediate chain (DIC) of the motor protein dynein thereby activating minus-end directed transport of membrane organelles along MT and regulating dynein activity by phosphorylation of the DIC component IC138 (Figure 3) (62, 228).
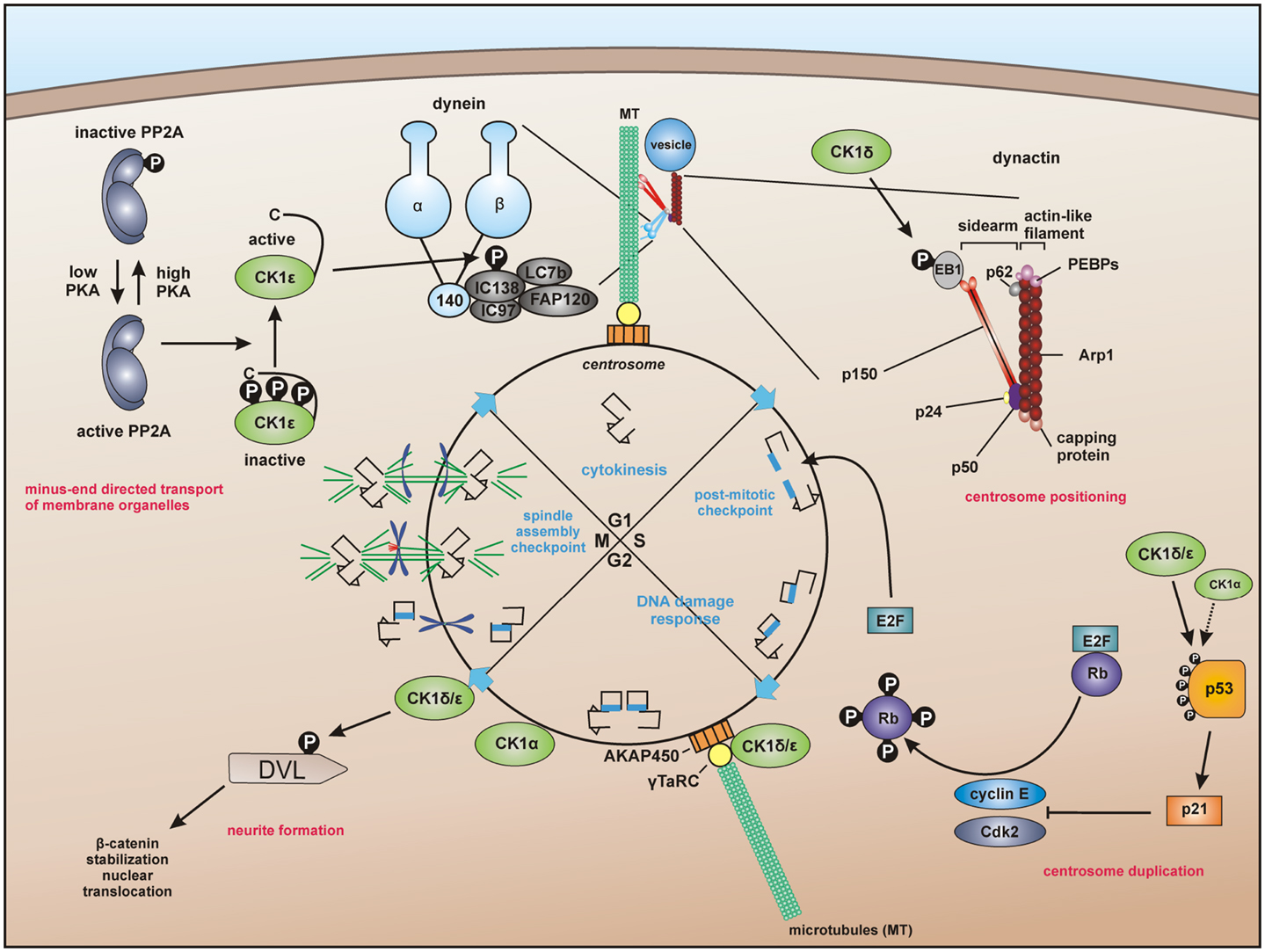
Figure 3. Centrosome-associated functions of CK1. For dynein-dependent transport along microtubules (MT), CK1ε phosphorylates the dynein intermediate chain (DIC) of dynein, likely IC138, thereby activating minus-end directed transport of membrane organelles along MT (62, 228). CK1δ and CK1ε are associated with the centrosome mediated through interaction with the scaffold protein AKAP450 (A-kinase anchor protein 450) (191, 193, 203). Both isoforms are related to Wnt-signaling and neurite outgrowth by phosphorylation of DVL (229, 230). In addition, CK1δ phosphorylates the end binding protein 1 (EB1), which is relevant for centrosome positioning during T-cell activation (74). Furthermore, a subpopulation of p53 in coordinated function with CK1 at the centrosome could ensure the integrity of the centrosome and thereby maintain genomic stability (231–233).
A particular interesting role of centrosome-associated CK1 has been proposed in regulating cell cycle progression by interaction with the Wnt pathway and p53 (Figure 3). CK1δ is associated to the centrosome and related to Wnt3-dependent neurite outgrowth. In this context, phosphorylation of DVL by centrosome-associated CK1δ facilitates neurite formation (32, 193, 229). CK1δ co-localizes with DVL2 at basal bodies and gradually accumulates at centrosomes when cells proceed through the cell cycle (230). The hypothesis of CK1 fulfilling regulatory roles at the centrosome is further underlined by the already discussed findings that CK1δ and ε are anchored at the centrosome through interaction with AKAP450 (see Regulation of CK1 Activity) (191) and that CK1δ phosphorylates EB1, which is relevant for centrosome positioning during T-cell activation (see CK1 in Immune Response and Inflammation) (74). Remarkably, further studies revealed that a subpopulation of p53 is located at the centrosome in order to prevent genomic instability. Therefore, the coordinated function of both CK1 and p53 could ensure the integrity of the centrosome and thereby maintain genomic stability (231–233).
CK1 in DNA Damage-Related Signal Transduction
CK1 family members can be considered as central components within the regulation of several cellular functions linked to DNA-processing or DNA damage [reviewed in Knippschild et al. (219)]. In context of DNA damage-associated signal transduction, p53 is activated initiating the activation of pathways ensuring centrosome integrity and genomic stability. This signaling network essentially involves coordinated action of CK1 and p53 (187, 231–233).
CK1α, δ, and ε are able to phosphorylate certain N-terminal target sites of p53 (Ser-6, Ser-9, Ser-15, Thr-18, and Ser-20) (187, 234–237). By phosphorylation of p53 (mostly at Ser-15 and Thr-18) CK1δ and ε decrease p53 binding affinity to its cellular counterpart Mouse double-minute 2 homolog (MDM2) resulting in increased levels of MDM2-released, active p53 (234, 238, 239). Conversely, phosphorylation of MDM2 at several serine residues within its central acidic domain (Ser-240, Ser-242, Ser-246, and Ser-383) results in increased MDM2-p53-binding and subsequent degradation of p53 under non-stress conditions. Phosphorylation of Ser-118 and Ser-121 by CK1δ, however, can mark MDM2 for SCF/β-TrCP (Skp1, Cullins, F-box/β-transducin repeat containing E3 ubiquitin protein ligase) binding and ubiquitination, finally leading to proteasomal degradation of MDM2 (Figure 4) (155, 239–241). Under normal conditions, CK1α has furthermore been suggested to be a key player promoting p53 inhibition and degradation by MDM2. Therefore, CK1α is physically interacting with MDM2 resulting in p53 degradation. Inhibition or depletion of CK1α as well as inhibition of CK1α-MDM2 association leads to p53 stabilization (208, 242). For the MDM2 homolog MDMX, phosphorylation of Ser-289 by CK1α has been confirmed resulting in increased binding to p53 and subsequent inhibition of p53 transcriptional function (156).
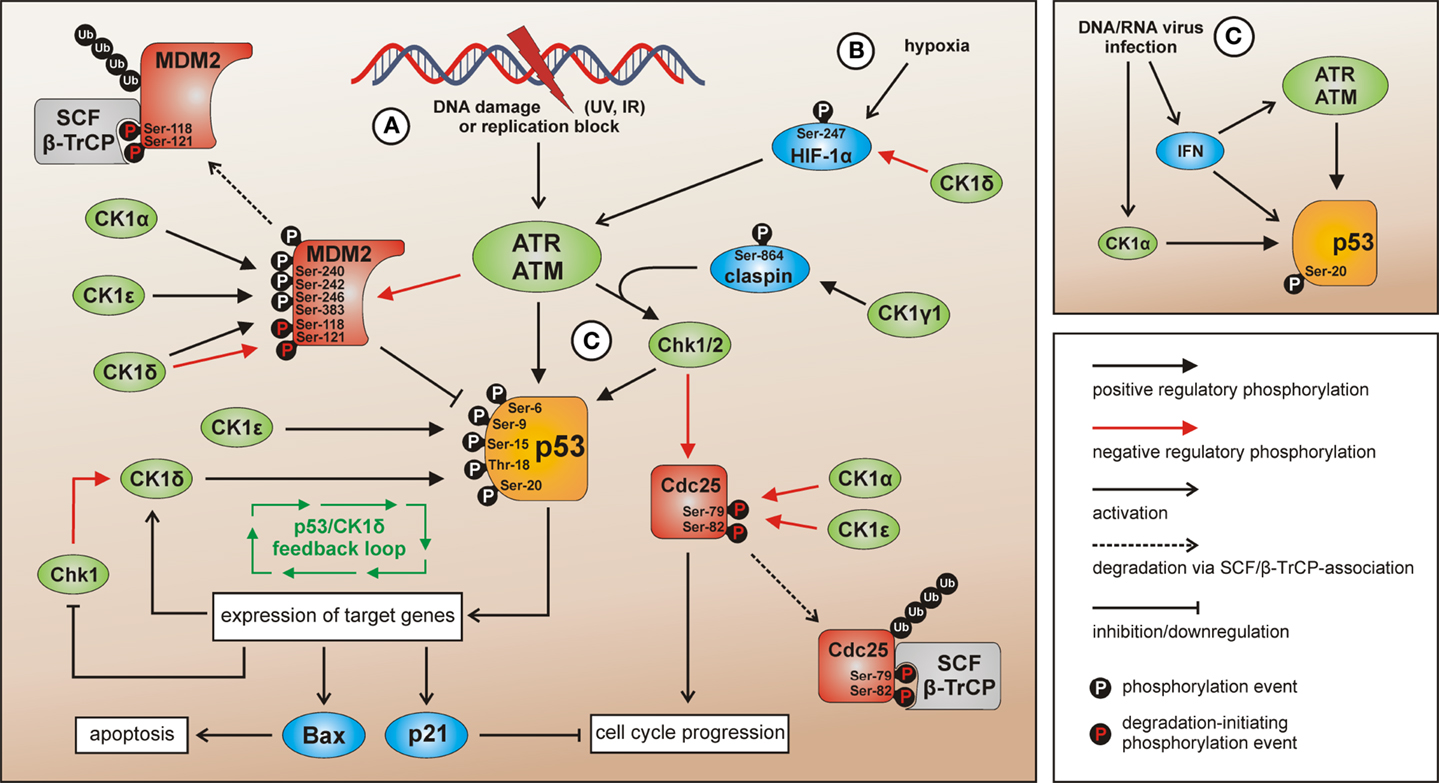
Figure 4. CK1 isoforms in DNA damage-induced signal transduction. After induction of DNA damage (situation A) p53 and Chk1/2 are activated by ATR/ATM-mediated phosphorylation while the p53-regulatory component MDM2 is inhibited. The activation of Chk1 is supported by claspin whereas Chk1/claspin-binding is promoted by CK1γ1-mediated phosphorylation of claspin (162). The CK1 isoforms α, δ, and ε are able to activate p53 by site-specific phosphorylation (187, 234, 235, 237). Activated p53 in turn induces the expression of target genes like Bax (leading to apoptosis), p21 (leading to cell cycle arrest), and also CK1δ (autoregulatory feedback loop) (187). MDM2-mediated degradation of p53 can be activated via interaction with and phosphorylation by CK1α, but also through phosphorylation by CK1δ or ε leading to enhanced binding of MDM2 to p53. CK1δ-mediated phosphorylation of Ser-118 and Ser-121 however marks MDM2 for proteasomal degradation (155, 239–241). In case Chk1/2 gets activated after DNA damage the phosphatase Cdc25, normally initiating cell cycle progression, is blocked by inhibitory phosphorylation and subsequent degradation. In the regulation of Cdc25 inhibition and degradation also CK1 isoforms α and ε are involved (141, 143). Signaling mediated by p53 can also be initiated by hypoxia (via CK1δ-regulated HIF-1α; situation B) (216, 243, 244) or DNA/RNA virus infection (via IFN and/or CK1α-related signal transduction; situation C) (236, 245). Depicted phosphorylation events refer to reported CK1-specific target sites.
Among the target genes activated by p53 following genotoxic stress also transcription of CK1δ can be induced (187). Given the previously discussed fact that p53 can be activated by CK1δ-mediated phosphorylation in this network, an autoregulatory feedback loop between CK1δ and p53 has been suggested (Figure 4).
Apart from DNA damage, p53 activation can also be induced by hypoxia. Herein, p53 levels are stabilized via HIF-1α and its positive regulatory effect on ATM/ATR (ataxia telangiectasia-mutated/ataxia telangiectasia and Rad3-related) (243, 244). As discussed previously, HIF-1α represents a substrate for CK1 and its transcriptional activity can be negatively regulated by CK1δ-mediated phosphorylation (216). However, since this modification has no effect on HIF-1α protein stability, the precise role of CK1δ-mediated HIF-1α phosphorylation in regulating ATM/ATR- and p53-specific functions under hypoxic conditions remains to be characterized.
More recent work suggested that interferon (IFN)-related signaling is able to activate p53 as a response to loss of epigenetic gene silencing (246). Among other critically involved epigenetic regulators, UHRF1 (ubiquitin-like, with PHD and RING finger domains 1) regulates the maintenance of DNA methylation during DNA replication (247). The stability of UHRF1 is regulated by proteasomal degradation including a priming step by CK1δ-mediated phosphorylation of Ser-108 thereby creating a recognition site for the SCF/β-TrCP ubiquitin ligase (104). Consequences of this negative regulatory connection between CK1δ and UHRF1 may also include the loss of stable DNA methylation and IFN-dependent activation of p53.
DNA/RNA virus infection has been described as a further mechanism resulting in p53 activation. This effect might be mediated via IFN-related p53 accumulation (245) but also via CK1-dependent signaling. In this context, CK1α-mediated phosphorylation of p53 at Ser-20 is induced after infection of T-cells with human Herpes virus 6B (HHV-6B). This phosphorylation event stabilizes the binding of p53 to the transcriptional co-activator p300. Therefore, CK1α takes part in gene regulation following virus infection induced p53 activation (236). Also infection with SV40 interferes with the p53 signaling network. SV40 large T-antigen (T-Ag) inactivates p53-dependent transcriptional activation whereas the oncogenic properties of T-Ag are enhanced by CK1-mediated phosphorylation (189, 190, 248). Moreover, as a consequence of SV40 infection/transformation, MDM2 is metabolically stabilized, post-translationally altered, and able to build trimeric complexes with T-Ag and p53 as well as complexes with free p53 thereby inhibiting proteasomal degradation of p53 (249).
Abnormalities in p53 are also related to phenotypes of premature aging. Recently, a mechanistic connection between the proteasome activator REGγ, CK1δ, and p53 has been demonstrated using a mouse model for premature aging. In this pathway, CK1δ is degraded after direct binding to REGγ. Subsequently, degradation of MDM2 is disturbed due to the lack of CK1δ and p53 levels decrease. These findings provide new insights to the conversely discussed anti- and pro-aging effects of p53 (250).
Obviously, CK1 family members are involved in p53-related signal transduction in response to cellular stress conditions in numerous ways (Figure 4). However, in most cases upstream regulators and the mechanism of CK1 activity regulation in these pathways still remain unknown. Another component in DNA damage-initiated signal transduction, being targeted by CK1 isoforms, is the protein phosphatase Cdc25A (cell division cycle 25A). Activation of cyclin-dependent kinases (Cdks) by dephosphorylation mediated by Cdc25 is required for cell cycle progression from G1 to S phase (251). Among phosphorylation by other cellular kinases, site-specific phosphorylation of Cdc25A by CK1α and ε at residues Ser-79 and Ser-82 targets Cdc25A for degradation via the ubiquitin-proteasome pathway (141, 143). This CK1-regulated degradation of Cdc25A supports DNA damage-induced cell cycle arrest, which is mediated via inhibition of Cdks by p53 and p21 (252). Since CK1 isoforms are involved in both, the degradation of Cdc25A as well as of p53, CK1 family members might act in a synergistic way to initiate cell cycle arrest.
In addition, CK1γ1 is related to DNA damage signaling by catalyzing the phosphorylation of claspin, an adaptor protein critically involved in ATR-mediated activation of Chk1. In this context, CK1γ1-mediated phosphorylation of claspin enhances its binding to Chk1 (162). Chk1 in turn has been identified as a cellular kinase phosphorylating CK1δ leading to decreased CK1δ-specific activity (38). The significance of this observation for the p53/MDM2/CK1-signaling network remains to be determined. However, given the information that Chk1 is down-regulated by p53 activation the Chk1/CK1δ/p53-interconnection might be involved in fine-tuning the negative regulatory effect of p53 on Chk1 (253).
In hematopoietic cells, the physical interaction of CK1ε with PTEN (phosphatase and tensin homolog) has been proposed to modulate cell survival. Normally, constitutively active Akt kinase or Akt activated by the upstream phosphatidylinositol 3-kinase (PI3K) leads to the inhibition of p53 and p53-induced apoptosis, thereby providing a resistance mechanism for genotoxic stress (254, 255). However, in case PTEN is stimulated as shown for the interaction of PTEN with CK1ε, PI3K-mediated Akt activation is inhibited. Subsequent inhibition of p53 via active Akt is circumvented and the cells’ sensitivity toward genotoxic stress can be restored (256).
In the context of Akt signaling, CK1α was reported to affect DEPTOR, an inhibitor of the mTOR (mammalian Target of Rapamycin) kinase, which regulates cell growth, proliferation, and survival [reviewed in Sarbassov et al. (257)]. Phosphorylation of DEPTOR by CK1α leads to βTrCP-mediated proteasomal degradation of DEPTOR resulting in activation of mTOR signaling, which is consistent with DEPTOR down-regulation and mTOR activation found in many cancers (258). Therefore, CK1α might provide a therapeutic target for the treatment of cancers characterized by low DEPTOR levels and activation of mTOR signaling, leading to increasing DEPTOR levels, and inhibition of mTOR signaling. Paradoxically, DEPTOR is overexpressed in multiple myeloma, which is necessary for PI3K-mediated activation of Akt and thereby inhibition of p53 and p53-induced apoptosis (259, 260).
Under conditions of genotoxic stress rapid changes in connexin-43 (Cx43) leading to alterations in gap junction-dependent intercellular communication have been observed in corneal endothelial cells associated with stabilization of gap junction communication (261). Earlier reports already showed phosphorylation of Cx43 by CK1δ, which stimulates the incorporation of Cx43 into gap junction plaques and which therefore most likely also takes part in long term cellular adaptations in response to genotoxic stress (76).
Further DNA-associated proteins being modulated by CK1 isoforms are topoisomerases. For topoisomerase IIα, phosphorylation of Ser-1106 by CK1δ and ε has been demonstrated (98). This phosphorylation event is linked to enhanced DNA-cleavage activity of topoisomerase IIα via the stabilization of topoisomerase-DNA cleavable complexes after etoposide treatment (98).
CK1 in Circadian Rhythm and Its Connections to Stress Response
In almost every higher organism, an autonomous timer is known and referred to as the circadian clock. This timer consists of a signal transduction pathway to integrate external signals for time adjustment, a molecular oscillator generating the circadian signal, and a signal transduction pathway controlling the circadian periodicity of certain biological processes. Therefore, circadian proteins are closely connected to key regulators of the cell cycle, oxidative stress, and carcinogenesis. Basically, in the mammalian circadian clock the positive regulators CLOCK and brain and muscle ARNT-like protein (BMAL1) as well as the negative regulators PERIOD (PER) and CRYPTOCHROME (CRY) form an oscillating system controlling their own expression levels (Figure 5) [reviewed in more detail in Kelleher et al. (262)].
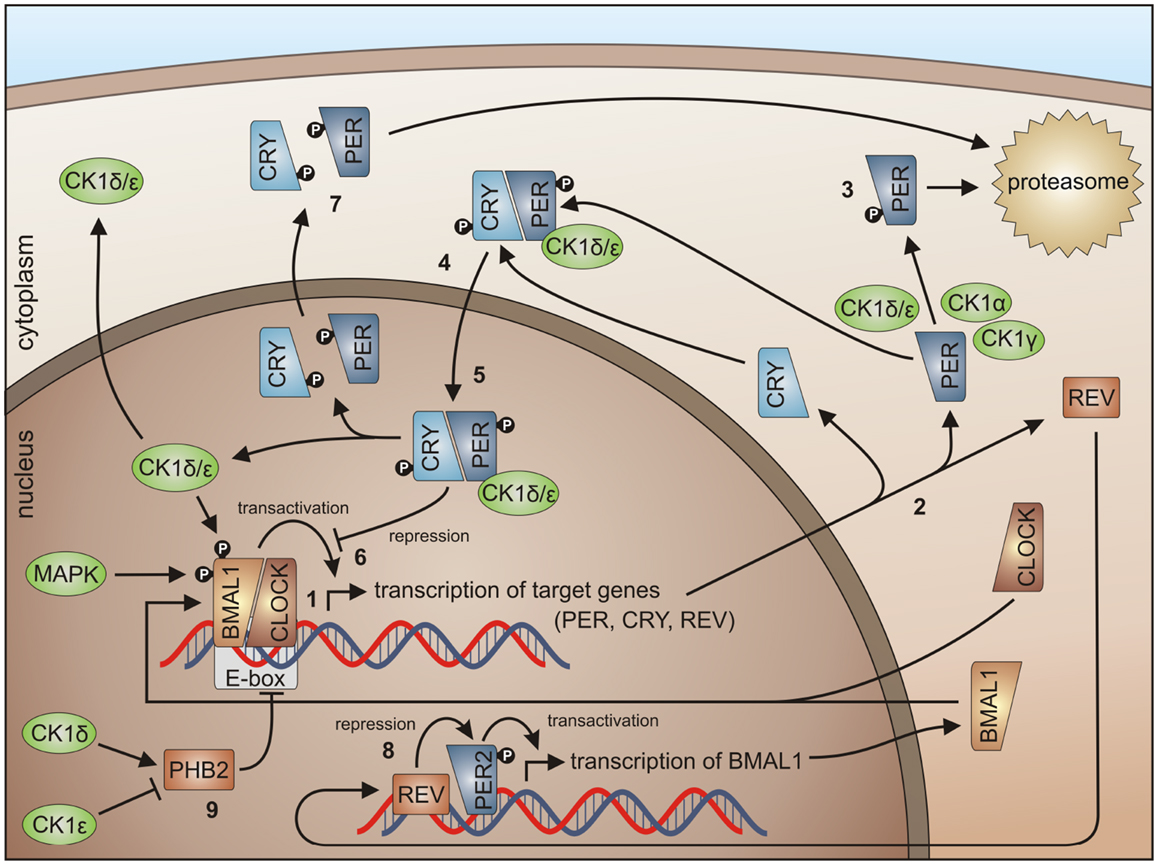
Figure 5. CK1 in circadian rhythm regulation. By binding of the BMAL1/CLOCK heterodimer to the E-box, transcription of E-box-containing genes is initiated (1) (here shown for PER1-3, CRY1 and 2, and REV-ERBα), the transcripts are translated in the cytoplasm (2). Degradation of cytoplasmic PERs is triggered mainly by CK1δ and ε, but also by isoforms α and γ (3), while PER degradation is inhibited by binding of CRYs to PERs (4). Subsequently, complexes of CRY/PER and CK1δ/ε translocate to the nucleus (5). In the nucleus the CRY/PER complex represses the transcriptional activation of BMAL1/CLOCK target genes (6). CRYs and PERs finally shuttle back to the cytoplasm for proteasomal degradation (7). Repression of BMAL1 expression by REV-ERBα represents a second negative feedback loop (8). Together, these feedback loops are able to generate cyclic expression of BMAL1 and E-box-containing genes [for review see Knippschild et al. (219) and Cheong and Virshup (195)]. CK1δ and ε differentially effect expression of the period length modulator PHB2. Whereas CK1δ is able to promote PHB2 transcription, its expression is reduced by CK1ε (9) (8, 263).
Linking circadian rhythm to cell cycle control, the heterodimer CLOCK/BMAL1 transcriptionally controls the expression of cell cycle regulators. PER1 interacts with ATM and Chk2 (264), whereas TIM, the mammalian homolog of Drosophila timeless protein, interacts with Chk1, ATR, and the ATR-related protein ATRIP (265). Furthermore, BMAL1 was identified to be necessary for p53-dependent growth arrest in response to DNA damage (266). Within the metabolism of reactive oxygen species (ROS) circadian proteins also seem to be involved, since the circadian clock could offer reliable control of daily variation in antioxidant response necessary to counteract increased oxidative stress. This connection is reasonable and important as oxidative stress is linked to the pathogenesis of cardiovascular diseases, atherosclerosis, and cancer (267). As an example, BMAL1 deficiency leads to chronic oxidative stress and an accelerated aging phenotype in mice (268). Conversely, activity of the circadian clock itself can be regulated by components of ROS metabolism (269). Finally, the circadian clock is also linked to the development of cancer. For PER2 mutant mice increased formation of radiation-induced lymphomas was reported and the frequency (FRQ) of intestinal and colonic polyps was increased in APCmin/+PER2m/m mice compared to APCmin/+ littermates (270).
In order to control the circadian rhythm involved, regulating components are subject to post-translational modifications like reversible phosphorylation (271). In general, CK1 isoforms δ and ε are able to phosphorylate and regulate the clock proteins BMAL1 and CRY and can modulate the expression of the period length modulator prohibitin 2 (PHB2) (114, 263). CK1δ is seen as an important regulator in circadian rhythm but also the involvement of other CK1 isoforms has been detected. CK1δ and ε are able to influence the length of the circadian period by regulating the stability and subcellular localization of PER (Figure 5) (149, 163, 272, 273). Phosphorylation of PER1 by CK1ε masks the nuclear localization signal of PER1 by conformational changes and marks PER for proteasomal degradation (149). CK1δ and ε interact with PER/CRY complexes thereby promoting nuclear localization of PER/CRY complexes (149, 274). In a high-throughput compound screening also CK1α was found to stimulate the degradation of PER1. In this screen, the protein kinases CK1α, CK1δ, and ERK2 were identified as targets for the compound longdaysin. However, CK1α binding affinity to PER1 is much weaker than for CK1δ or ε (275). The same is true for CK1γ (276). Thus, CK1δ and ε can be regarded as redundant for PER phosphorylation and essential for nuclear accumulation of PER (277). Inhibition of CK1δ and ε by the pan-CK1δ/ε inhibitor PF-670462 led to remarkably lengthened circadian rhythms (in vivo locomotor activity) and molecular oscillations (in vitro in the suprachiasmatic nucleus and peripheral tissue slices). These observations could not be made using the CK1ε-specific inhibitor PF-4800567 (278). PF-4800567 effectively blocked CK1ε-mediated nuclear localization of PER3 and degradation of PER2 but only showed minimal effect on the circadian clock in cycling Rat1 fibroblasts (273). The CK1ε tau mutation, however, which was discovered in the Syrian hamster as the first mammalian circadian mutation, was characterized as a gain of function mutation resulting in clock acceleration. In mice expressing the CK1ε tau mutation increased phosphorylation of PER1 and 2 can be detected leading to increased degradation of nuclear and cytoplasmic PER and acceleration of the mammalian clock (279, 280). Um and colleagues discovered, that the circadian period of Rat1 fibroblasts treated with the diabetes drug metformin was shortened by 1 h. By metformin treatment, AMP-activated kinase (AMPK) is activated, which phosphorylates CK1ε at Ser-389 leading to increased activity of CK1ε and subsequent degradation of Per2 (281). A higher level regulator of CK1ε activity in circadian rhythm is protein phosphatase 5 (PP5), which can raise the activity of CK1ε by dephosphorylation. As a consequence, phosphorylation by CK1ε and subsequent degradation of PER is also increased (282). Recently, CK1δ (but not CK1ε) has been shown to be crucial for the circadian timing mechanism in zebrafish (283).
Presented observations point to PER proteins as multikinase targets, which can be multiply phosphorylated and thereby regulated. Herein, the balance between phosphorylation and dephosphorylation by phosphatases is of certain importance. In cells deficient for CK1δ and ε, phosphorylation of PER is disturbed and PER proteins remain cytoplasmic. In case protein phosphatase 1 (PP1) is disrupted, phosphorylation of PER is accelerated. This effect is specific to PP1 and in contrast to previous Drosophila studies cannot be observed for PP2A (276).
CK1-Signaling in Apoptotic Pathways
For several CK1 isoforms, an involvement in the regulation of apoptotic signal transduction has been described. CK1α, δ, and ε are components of Fas-mediated apoptosis and induce an activation of initiator caspase 8. Here the pro-apoptotic protein Bid, which belongs to the Bcl-2 family, is of major interest. Amino acids Ser-64 and Ser-66 of Bid are supposed to be major targets for CK1-mediated phosphorylation while Thr-58 is targeted by CK2. Only unphosphorylated Bid can be processed by caspase 8-mediated proteolysis and can participate in cytochrome c-mediated apoptosis. Accordingly, inhibition of CK1 and CK2 induces accelerated Fas-triggered apoptosis by blocking inhibitory phosphorylation of Bid. Vice versa an overexpression of CK1ε and CK2 leads to a decreased number of apoptotic cells due to increased phosphorylation of Bid, blocking its caspase 8-mediated processing. Therefore, phosphorylation of Bid by CK1δ and ε and CK2 can inhibit Fas-mediated apoptosis (151).
Moreover, CK1 (isolated from pig spleen) can phosphorylate the p75 neurotrophin receptor, thereby negatively regulating p75-mediated apoptosis (284). CK1α is involved in apoptosis by interaction and phosphorylation of retinoid X receptor (RXR), a class of retinoic acid receptors regulating cell survival by building heterodimers with NGF1B (nerve growth factor 1B), IGFBP-3 (insulin-like growth factor binding protein 3), and β-catenin. In this case, CK1 activity inhibits the induction of apoptosis by RXR agonists (85, 285–287).
Furthermore, CK1α is able to phosphorylate FADD (Fas-associated protein with death domain) at Ser-194 in vitro as well as in vivo and is supposed to be involved in regulating non-apoptotic functions of FADD like cell cycle interaction, sensitivity toward chemotherapeutics, and nuclear localization (176, 288). In erythrocytes, CK1α modulates cytosolic calcium activity and thereby regulates programed cell death (289).
Participation of CK1 in the Development of Cancer
During animal development, a precise coordination of cell patterning events is required to ensure appropriate organ architecture and size. Several developmental pathways control growth, proliferation, and apoptosis by strict regulation, which can result in pathological conditions when dysregulated. The Wnt (Wingless/Int-1), Hh (Hedgehog), and Hippo signaling pathways are important in tissue development, growth, and homeostasis (290–293). Aberrant activation of these pathways as well as mutations of components of these pathways has been linked to various cancer entities (294–298). Due to the contribution of CK1 family members in pathways associated with growth and development, the following sections concentrate on the current knowledge of CK1 participation and regulation in the Wnt, Hh, and Hippo signaling pathways.
CK1 and the Wnt Pathway
Components of the Wnt-signaling pathway are involved in many developmental processes including dorsal axis formation, tissue patterning, and establishment of cell polarity (299–302). In addition, Wnt/β-catenin-mediated signaling plays an important regulatory role in cell proliferation processes in both, embryonic and mature organisms. Mutations in Wnt pathway components have been found in various human cancers, including cancers of the skin, liver, brain, and colon (291, 303–312).
In the canonical Wnt/β-catenin signaling pathway, all CK1 family members are involved. However, this involvement is quite complex. So far, positive as well as negative regulatory functions have been described. In absence of the Wnt ligand CK1α interacts with and phosphorylates Axin, adenomatous polyposis coli (APC), and β-catenin (at Ser-45), thereby priming β-catenin for further phosphorylation by GSK3β and subsequent degradation (195, 313) (Figure 6A). After binding of Wnt ligand to Frizzled (Fzd) the Wnt co-receptor LRP5/6 is phosphorylated either by membrane-bound CK1γ (positive regulation) (86) or by CK1ε (negative regulation) (314). Phosphorylated LRP5/6 then recruits Axin and the β-catenin destruction complex to the membrane and inhibits GSK3β. Wnt-activated CK1δ and CK1ε phosphorylate Axin as well as the scaffold protein DVL at multiple sites and can introduce a conformational change to the β-catenin destruction complex followed by dissociation of several components, thereby preventing β-catenin from being phosphorylated and degraded (148, 195). Recently, RNA helicase DDX3 was identified as a regulatory subunit of CK1ε in Wnt-signaling. Wnt-activation promotes recruitment of DDX3 to CK1ε and binding directly stimulates kinase activity, promoting phosphorylation of DVL, finally leading to stabilization of β-catenin (103). Accumulated β-catenin then translocates to the nucleus to activate the expression of TCF/LEF (T cell factor/lymphoid enhancing factor)-triggered target genes (Figure 6B) (291). CK1ε is also involved in the formation of an active transcription complex by phosphorylating TCF3 thereby mediating its activation and binding to β-catenin (113).
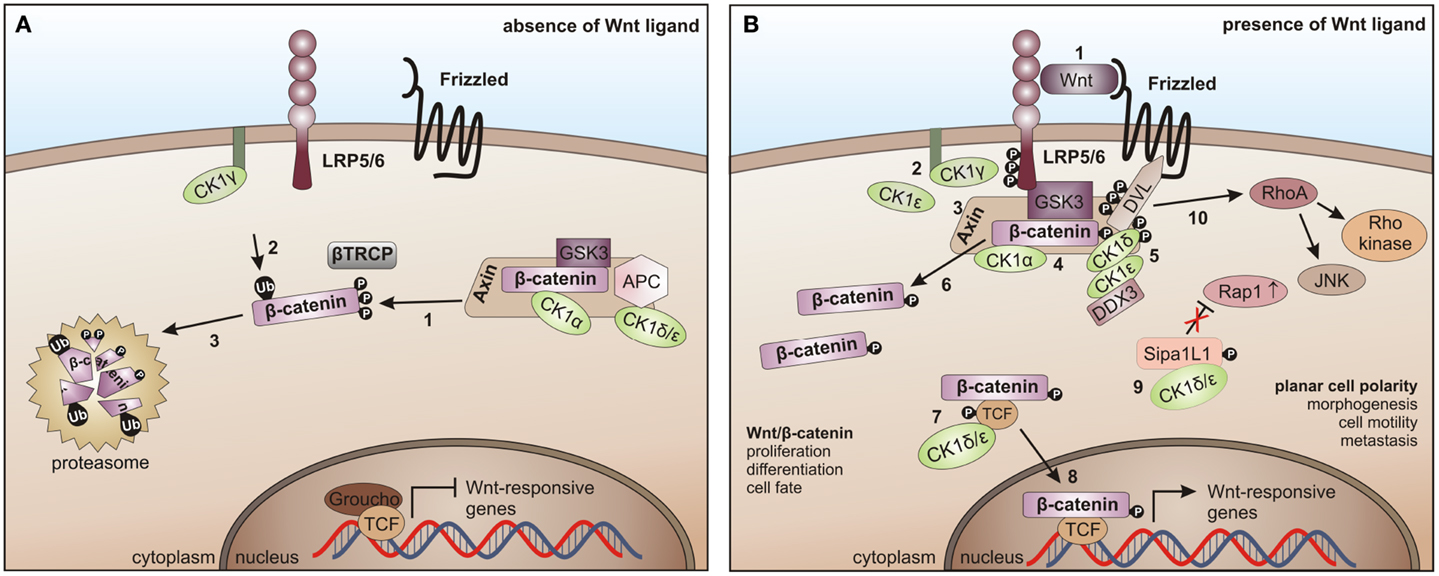
Figure 6. CK1 in Wnt-signaling. (A) In the absence of the Wnt ligand, β-catenin is progressively phosphorylated by CK1α and GSK3 (1), recruited to β-TrCP for ubiquitination (2), and thereby primed for proteasome- dependent degradation (3). (B) After binding of Wnt to Frizzled and LRP5/6 (1), LRP5/6 is phosphorylated by CK1γ (positive regulation) and CK1ε (negative regulation) (2). It then recruits Axin and the β-catenin destruction complex to the membrane and inhibits GSK3 (3, 4). Wnt-activated CK1δ and ε phosphorylate Disheveled (DVL) and Axin (5), induce a conformational change in the β-catenin destruction complex and initiate the dissociation of various components (6). CK1ε cooperates with DDX3 in phosphorylating DVL (7). Also, TCF3 can be phosphorylated by CK1δ and ε thereby increasing its binding to β-catenin followed by the nuclear translocation of TCF3/β- catenin (8). The non-canonical Wnt pathway is positively regulated by CK1δ- and ε-dependent release of Rap1 from Sipa1L1 inhibition (9). The Rho/JNK signaling cascade is activated after phosphorylation of DVL (10) [adapted from Cheong and Virshup (195)].
CK1ε is associated with a positive regulatory function by joining the Wnt multi-protein complex to phosphorylate DVL, which in turn gets activated and inhibits GSK3β, finally resulting in stabilization of β-catenin (148).
Signaling in the non-canonical Wnt pathway is positively regulated by CK1δ and ε, which release Rap1 from Sipa1L1 inhibition. Subsequent to phosphorylation of DVL the Rho/JNK signaling cascade can be activated (Figure 6B) [reviewed in Cheong and Virshup (195)].
CK1 in the Hedgehog Pathway
The Hh signaling pathway regulates a variety of processes during embryonic development such as differentiation, proliferation, and organogenesis (290). In the adult organism, Hh signaling is significantly reduced but plays a critical role in regulating epithelial maintenance and regeneration of organs, which undergo constant renewal; among them, epithelia of internal organs and brain (315). Therefore, mutations or dysregulation of components of this pathway are associated with tumorigenesis and cancer development, including basal cell carcinomas, medulloblastomas, gliomas, gastrointestinal tumors, prostate cancer, and hematological malignancies (315–318).
In mammals, major components of the Hh pathway are represented by the three Hh homologous ligands Sonic hedgehog (Shh), Indian hedgehog (Ihh), and Desert hedgehog (Dhh), the negative regulatory 12-pass membrane receptor Patched (PTCH), the positive regulatory 7-pass membrane protein smoothened (SMO), the glioma-associated oncogene (GLI) transcription factors GLI1, GLI2, GLI3, a multi-protein complex consisting of intraflagellar transport proteins, protein kinase A (PKA), GSK3, CK1, and suppressor of fused (SUFU) (319). In absence of Hh ligands, PTCH inhibits the localization of SMO to the cilia cell surface and represses SMO activity, thereby suppressing signal transmission via the GLI transcription factors into the nucleus. PKA, GSK3β, and CK1 phosphorylate the GLI transcription factors leading to their proteolytic processing into the repressor forms, which cannot activate target gene transcription (Figure 7A) (320, 321). Hh signal transduction is initiated upon binding of a Hh ligand to PTCH, thereby releasing SMO from PTCH-mediated inhibition, leading to its accumulation on cilia cell surface and consequent activation and release of the GLI transcription factors from the multi-protein complex. Activated GLIs then translocate to the nucleus, where they induce transcription of Hh target genes (Figure 7B) (319).
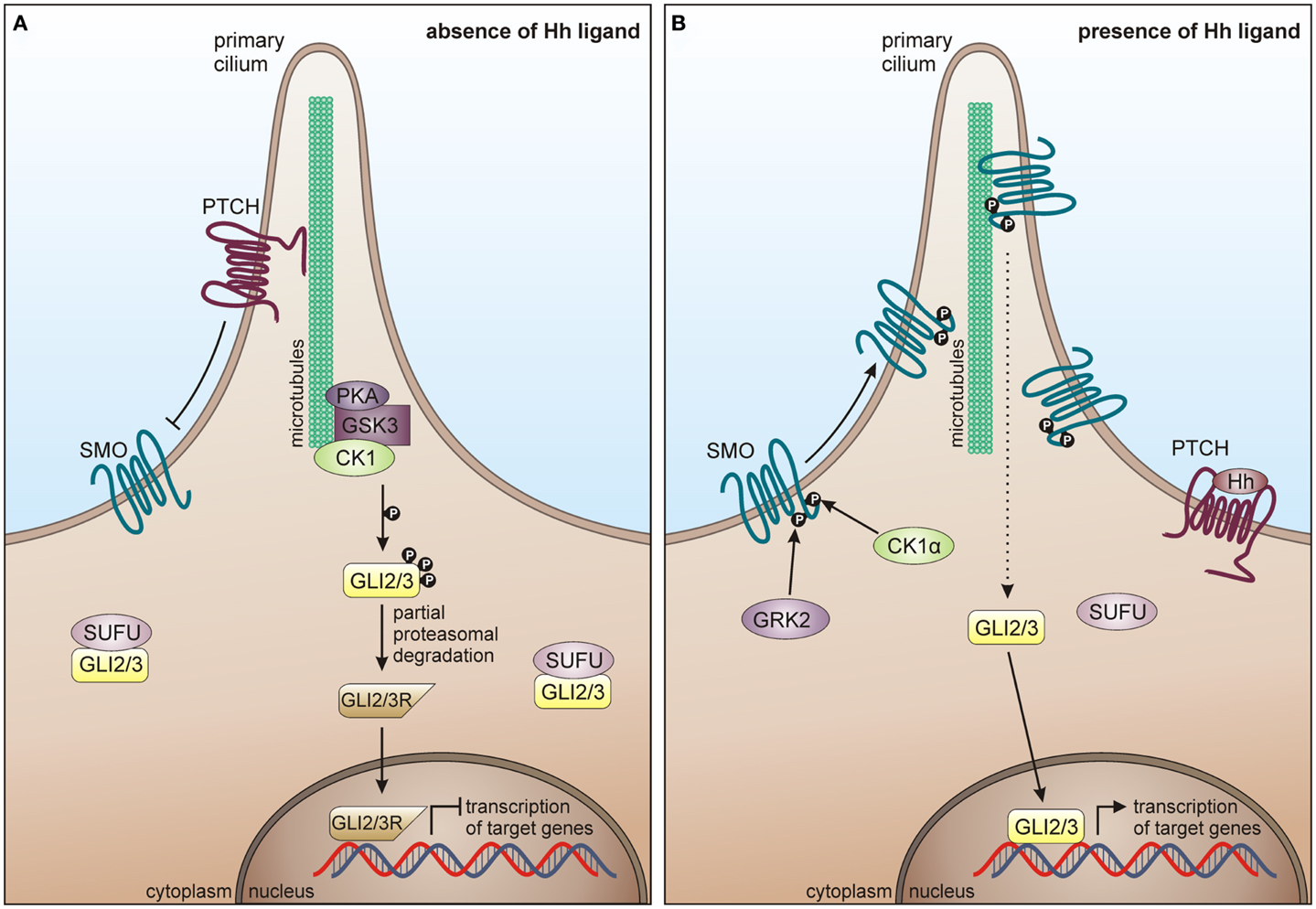
Figure 7. CK1 in Hh signaling. (A) In absence of Hh ligand, PTCH localizes in the cilium and inhibits surface trafficking and cilia localization of SMO. GLI proteins are phosphorylated by PKA, GSK3β, and CK1, which lead to proteasome-dependent cleavage of GLI into a N-terminally truncated form, generating the repressor forms GLI2R and GLI3R. GLI2/3R translocate to the nucleus and inhibit translation of Hh target genes. Furthermore, SUFU prevents GLI from activating Hh target genes, by binding it in the cytoplasm and the nucleus. (B) In response to Hh, SMO is activated by GRK2 and CK1α-dependent phosphorylation and enters the primary cilium. Activated SMO orchestrates a signaling cascade, eventually resulting in the dissociation of the SUFU-GLI complex and the translocation of full-length GLI2/3 to the nucleus, where Hh target gene expression is induced.
In 2002, Price and Kalderon postulated a negative regulatory role of CK1 in Hh signaling in Drosophila melanogaster (322). They demonstrated that CK1δ- and GSK3-mediated phosphorylation of Ci-155 (full-length Cubitus interruptus, the Drosophila homolog of GLI2 and GLI3) at PKA primed sites is required for the partial proteolysis of the transcription factors, thereby preventing Hh target gene transcription [reviewed in Price (323)]. The PKA, GSK3, and CK1 sites are conserved in Ci, GLI2, and GLI3, which are all similarly processed and may play similar roles in Drosophila and vertebrates (324–327). Furthermore, Wang and Li demonstrated, that CK1 and GSK3 phosphorylation sites are needed to process GLI3 (327). CK1 has also been implicated in positive regulation of SMO. Chen and co-workers demonstrated that mammalian SMO is activated via multiple phosphorylation events mediated by CK1α and G protein coupled receptor kinase 2 (GRK2), thereby inducing its cilia accumulation and active conformation (328).
CK1 in the Hippo Pathway
During development, the evolutionary conserved Hippo pathway contributes to several processes, which restrict organ size by controlling cell proliferation and apoptosis [reviewed in Zhao et al. (124)]. Consequently, pathway deregulation can trigger tumorigenesis and occurs in a broad range of human cancers. Abnormal Hippo activity is associated with cancer cell proliferation, enhanced cell survival, and maintenance of a stem cell phenotype [reviewed in Harvey et al. (329)].
The mammalian Hippo pathway is initiated by various growth suppressive signals like cell contact inhibition. The upstream kinases mammalian STE20-like protein kinase 1/2 (MST1/2), together with the scaffold proteins vertebrate homolog of Drosophila Salvador (WW45) and MOB kinase activator 1A/B (MOB1A/B) phosphorylate the large tumor suppressor 1 and 2 (LATS1/2). LATS1/2-dependent phosphorylation of the transcriptional co-activator Yes-associated protein (YAP) and its paralog Tafazzin (TAZ) then leads to YAP/TAZ inhibition by spatial separation from its nuclear target transcription factors TEAD (TEA domain) and SMAD (SMA/mothers against decapentaplegic) and additionally by phosphodegron-mediated degradation, preventing Hippo target gene transcription (Figure 8A) (330, 331).
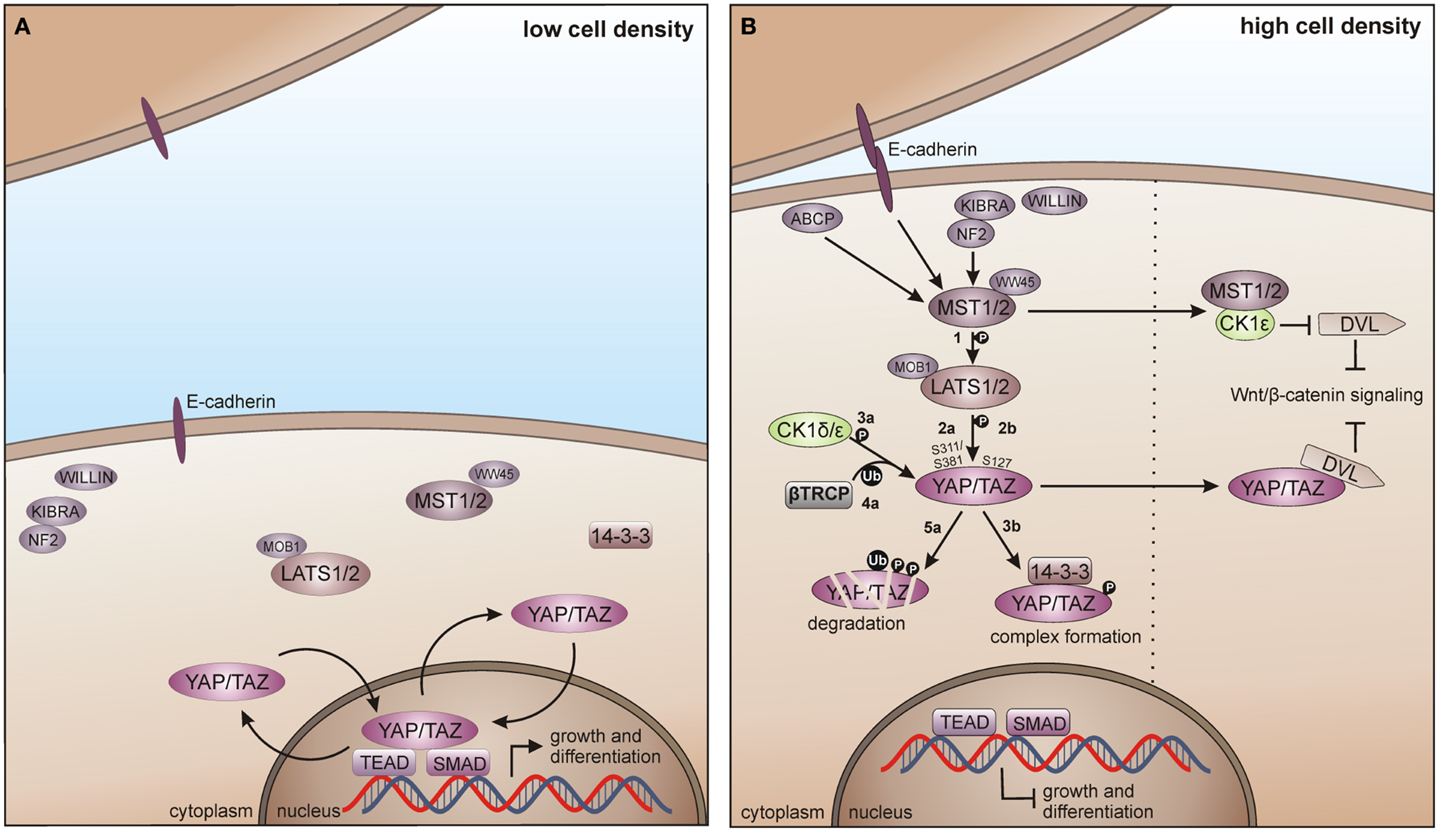
Figure 8. CK1 in Hippo signaling in vertebrates. (A) In absence of growth suppressive signals YAP/TAZ promotes tissue growth and differentiation by regulating the activity of different transcription factors in the nucleus, including SMADs and TEADs. (B) Cell-density activated pathway regulation is controlled by multiple upstream branches by activating the core kinase cassette that represses YAP/TAZ driven gene transcription, either by degradation of TAP/TAZ or by forming physical complexes, preventing its nuclear access. Initially, MST1/2 is activated by various components and phosphorylates LATS1/2 (1), which in turn phosphorylates TAP/TAZ on Ser-311 or Ser-381 (2a). This phosphorylation primes YAP/TAZ for further phosphorylation by CK1δ/ε (3a) and consequent recruitment of and ubiquitination by β-TrCP (4a), priming YAP/TAZ for degradation (5a). However, LATS1/2 driven phosphorylation of TAP/TAZ on Ser-127 (2b) leads to the formation of 14-3-3-YAP/TAZ complexes, which accumulate in the cytoplasm preventing YAP/TAZ access to the nucleus (3b). Hippo pathway regulates Wnt/β-catenin signaling by inhibition of DVL, either by MST1/2-mediated prevention of CK1ε-dependent phosphorylation of DVL, or by direct inhibition of DVL by YAP/TAZ. ABCP: apicobasal cell polarity protein.
Zhao and co-workers as well as Liu and co-workers identified CK1δ and ε as new temporal regulators of the Hippo pathway. YAP is phosphorylated by LATS on Ser-381 and this phosphorylation provides the priming signal for CK1δ or ε to phosphorylate a phosphodegron in YAP, which in turn recruits β-TrCP leading to YAP ubiquitination and degradation (124). Furthermore, TAZ phosphorylation at Ser-311 by LATS also leads to subsequent CK1ε-mediated phosphorylation of a phosphodegron in TAZ and consequently to its degradation (123). Xu and co-workers recently postulated the interaction of the Hippo and Wnt pathway via CK1ε. Herein, the Hippo upstream kinase MST1 is able to suppress the Wnt/β-catenin pathway by directly binding CK1ε, thereby preventing phosphorylation of DVL (Figure 8B) (332).
CK1-Related Tumorigenic Functions
The important role of CK1 family members within various signaling pathways is furthermore supported by reports linking CK1 isoforms to modulation of key regulatory proteins such as p53, MDM2, and β-catenin, which act as signal integration molecules in stress situations and generally can be seen as a key regulatory connection to tumorigenesis [for more detailed review see Knippschild et al. (219, 333), and Cheong and Virshup (195)]. Considering the importance of signals mediated by CK1δ and ε to finally ensure genome stability, it is obvious that mutations leading to changes in the activity or expression levels of CK1 isoforms or mutations of CK1-specific target sites in its substrates can contribute to the development of cancer (Table 2). Foldynová-Trantírková and co-workers provided evidence that mutations in CK1ε, which are frequently found in breast cancer, lead to loss of function in the Wnt/β-catenin pathway but result in activation of the Wnt/Rac1/JNK and Wnt/Ca2+ pathway, consequently leading to increased migratory capacity and decreased cell-adhesion (334). A mutation within the C-terminal region of CK1δ detected in an adenomatous colorectal polyp leads to a higher oncogenic potential and promotes the development of adenomas in the intestinal mucosa (335). Furthermore, conditional knock-out of CK1α in the intestinal epithelium leads to activation of p53 and Wnt-signaling, while in p53 deficient gut, loss of heterozygosity of the CK1α gene causes a highly invasive carcinoma, indicating that CK1α acts as a tumor suppressor when p53 is inactivated (336).
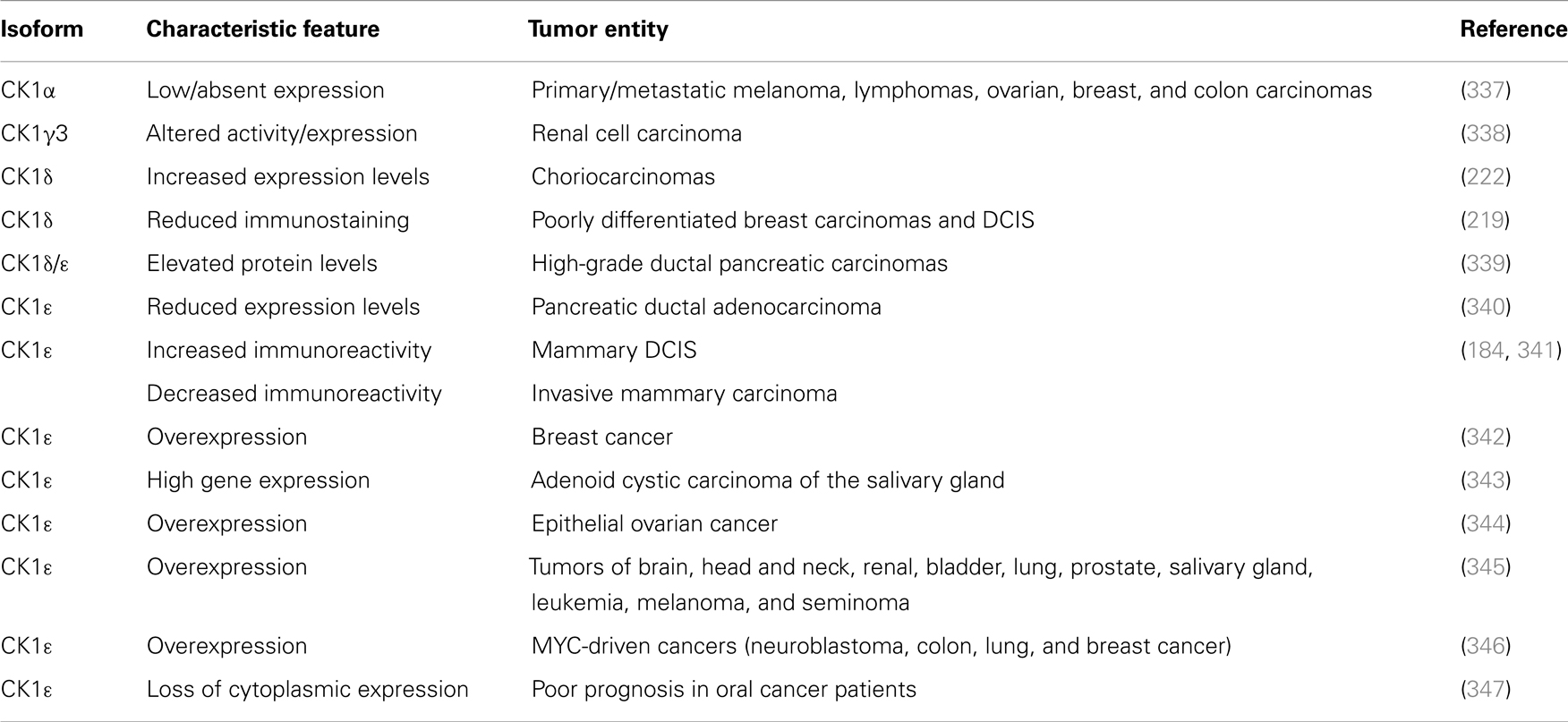
In 1981, Elias and co-workers already reported an increased nuclear CK1 kinase activity in AML patients (186). Until now, several reports link altered CK1 expression and/or activity to cancer. Reduced CK1α protein and mRNA expression levels in primary melanomas and melanoma metastases compared to benign melanocytic lesions or early-stage melanomas have been detected. In the same study, reduced CK1α expression was also observed in lymphomas, ovarian, breast, and colon carcinomas, compared with the respective benign tissue (337). In renal cell carcinoma elevated CK1γ3 expression and activity levels have been described (338), whereas in choriocarcinomas strong expression levels of CK1δ were detected (222). Changes in the immunoreactivity of CK1δ have been observed in breast carcinomas, depending on the grade of tumor differentiation. High-grade ductal carcinomas in situ (DCIS) as well as invasive poorly differentiated carcinomas show reduced CK1δ immunostaining, whereas well differentiated carcinomas and low grade DCIS show strong staining of tumor cells (219). Regarding CK1ε, Fuja and co-workers observed similar correlations between tumor differentiation and immunohistological staining (341). Expression of CK1ε is also down-regulated in mammary cancers in SV40-transgenic mice expressing SV40 T-Ag (184). A recent study suggests that CK1ε is overexpressed in breast tumors and acts as a pivotal regulator of mRNA translation and cell proliferation. CK1ε phosphorylates the negative-acting factor 4E-BP1 (eukaryotic translation initiation factor 4E binding protein 1), thereby preventing its inhibitory function on the translation initiation complex elF4E (eukaryotic initiation factor 4E) and consequently leading to dysregulated mRNA translation and breast cancer cell growth (342). Elevated protein levels of CK1δ and ε were also observed in single tumor cells of grade 3 tumors of ductal pancreatic carcinomas and inhibition of CK1δ and ε by the CK1-specific inhibitor IC261 reduced pancreatic tumor cell growth in xenografts (339). In contrast, Relles and co-workers detected reduced expression levels in pancreatic ductal adenocarcinomas (340). CK1ε expression is increased in adenoid cystic carcinomas of the salivary gland (343), in epithelial ovarian cancer (344), in tumors of brain, head and neck, renal, bladder, lung, prostate, and salivary gland, in leukemia, melanoma, and seminoma (345). Toyoshima and co-workers found that CK1ε expression is significantly correlated with MYCN amplification in neuroblastoma and poor prognosis. In addition, CK1ε expression has been associated with c-MYC in several other tumors such as colon, lung, and breast cancer (346). In a recent study, Lin and co-workers demonstrated that loss of cytoplasmatic CK1ε expression correlates with poor survival rates in oral squamous cell carcinoma (347). Järas and co-workers recently found that CK1α is essential for AML cell survival and treatment with the CK1-specific inhibitor D4476 results in highly selective killing of leukemia stem cells by reducing Rsp6 (radial spoke protein 6) phosphorylation and activation of p53 (Table 4) (348).
In summary, the data reported so far provide evidence that CK1 isoforms exhibit oncogenic features by promoting proliferation, genome instability, and inhibition of apoptotic processes. This assumption is also supported by the fact that CK1 isoforms are often overexpressed in tumors and that overexpression of CK1ε correlates with poor survival as shown for patients with ovarian cancer (344). However, this finding cannot be generalized and might depend on additional factors, as in the case of oral squamous cell carcinoma loss of CK1ε expression correlates with poor survival rates (347). In addition, the functions of CK1α in tumorigenesis are manifold making it difficult to classify it as oncogene or tumor suppressor. In AML CK1α seems to exhibit oncogenic features (348), whereas in intestinal epithelium loss of heterozygosity of the CK1α gene causes a highly invasive carcinoma, indicating that CK1α acts as a tumor suppressor when p53 is inactivated (336).
CK1 in Metastatic Processes
In many cases, CK1 family members can also be involved in the regulation of metastatic processes. However, their potential to promote or suppress metastasis seems to depend not on the specific isoform but on their position in cellular signal transduction and the cellular context. Phosphorylation of nm23-H1 by CK1δ and ε has been shown to induce complex formation of nm23-H1 with a cellular partner called h-prune. Both proteins are linked to proliferative disorders and the nm23-H1-h-prune complex formation has even been proposed to positively influence cell motility (349). With this link of CK1 kinase activity to nm23-H1-h-prune complex formation an obvious role for CK1 in the mediation of metastasis has been established (153).
Quite recently, the stability of metastasis-related proteins has been shown to be regulated by CK1δ-mediated phosphorylation. First, the epigenetic sensor UHRF1 is critically involved in the maintenance of DNA methylation patterns during DNA replication and can be linked to carcinogenesis and metastasis if dysregulated (350, 351). Second, as mentioned before, proteasomal degradation of UHRF1 is regulated by CK1δ-mediated phosphorylation (104). Similar findings have been reported for metastasis suppressor 1 (MTSS1, also known as MIM, missing in metastasis), an anti-metastatic protein whose degradation also is triggered by CK1δ-mediated phosphorylation at Ser-322, thereby inducing its interaction with SCF/β-TrCP (77, 352).
Furthermore, current reports demonstrate the involvement of CK1α in regulating the stability of metastasis-associated factors. When cell motility is induced the Rap guanine exchange factor (RAPGEF2) is phosphorylated by IKKβ and CK1α, initiating SCF/β-TrCP-mediated degradation. RAPGEF2 degradation-failure leads to inhibition of hepatocyte growth factor (HGF)-induced cell migration and expression of non-degradable RAPGEF2 suppressed metastasis of human breast cancer cells (165).
In canonical Wnt-signaling, CK1α has been positioned to be a tumor suppressor and cancer cells may activate proliferative processes via the Wnt/β-catenin pathway by suppressing CK1α expression. In the absence of CK1α, p53 is critically involved in controlling invasiveness as shown in a model for colon cancer (336). Re-expression of CK1α in metastatic melanoma cells reduced growth in vitro and metastasis formation in vivo (337). Consistent with these findings phosphorylation of β-catenin at Ser-45 by CK1α via activation by Wnt-5a has been shown to increase complex formation of β-catenin with E-cadherin thereby maintaining intercellular adhesion. Loss of Wnt-5a is thought to be associated with initial metastatic de-adhesion events (353, 354). Conversely, E-cadherin-mediated cell–cell contacts can be negatively regulated by CK1ε-mediated phosphorylation of E-cadherin at Ser-846 (69). In this context also, the Zn-finger transcription factor Snail is important as it can promote epithelial to mesenchymal transition (EMT) by down-regulating E-cadherin expression (355). Herein, CK1ε primes Snail for GSK3β-mediated phosphorylation, which marks Snail for degradation. Therefore, loss of CK1ε kinase activity prevents GSK3β-mediated phosphorylation and degradation of Snail supporting EMT and metastatic processes (122).
CK1-Specific Inhibitors
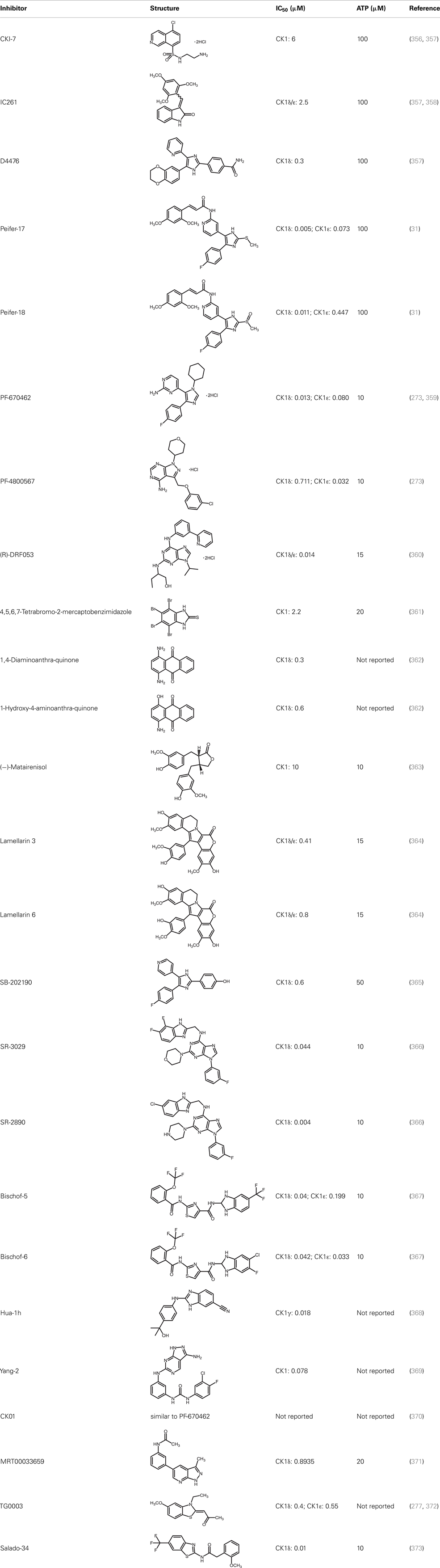
Due to the obvious involvement of CK1 isoforms in the pathogenesis of inflammatory and proliferative diseases and its contribution to the development of neuro-degenerative disorders, CK1 family members are attracting more and more attention as drug targets in regard to therapeutic applications. So far, several highly potent CK1-specific small molecule inhibitors have been identified (Table 3) and some have already been characterized for their therapeutic potential in animal models (Table 4). Most of these compounds are ATP-competitive type I inhibitors raising the problem of comparability of their effectiveness since their IC50 values have been determined at different ATP concentrations (see Table 3).
CKI-7 (N-(2-aminoethyl)-5-chloroisoquinoline-8-sulfonami de), was the first ATP-competitive inhibitor being described to show selectivity toward CK1 (356). Later, IC261 (3-[(2,4,6-trimethoxyphenyl)-methylidenyl]-indolin-2-one) and D4476 (4-[4-(2,3-dihydro-benzo)[1,4]dioxin-6-yl)-5-pyridin-2-yl-1H-imi dazol-2-yl]-benzamide) have been described as more potent and selective inhibitors, which also bind to the ATP binding pocket of CK1 (357, 358). Several effects reported for IC261-treated cells may however not be related to the selective inhibition of CK1 (380, 381). IC261 is also able to bind MT thereby inhibiting their polymerization similar to the spindle poison colchicine (380). Nevertheless, IC261 inhibits site-specific phosphorylation of p53 and Bid thereby inducing apoptosis in so-called type II cells (151, 187). Furthermore, its therapeutic potential has been demonstrated in xenotransplantation models for pancreatic cancer and neuroblastoma tumors (339, 346) (Table 4). However, it is still questionable whether the described anti-tumorigenic effects of IC261 are all mediated through selective inhibition of CK1δ and ε.
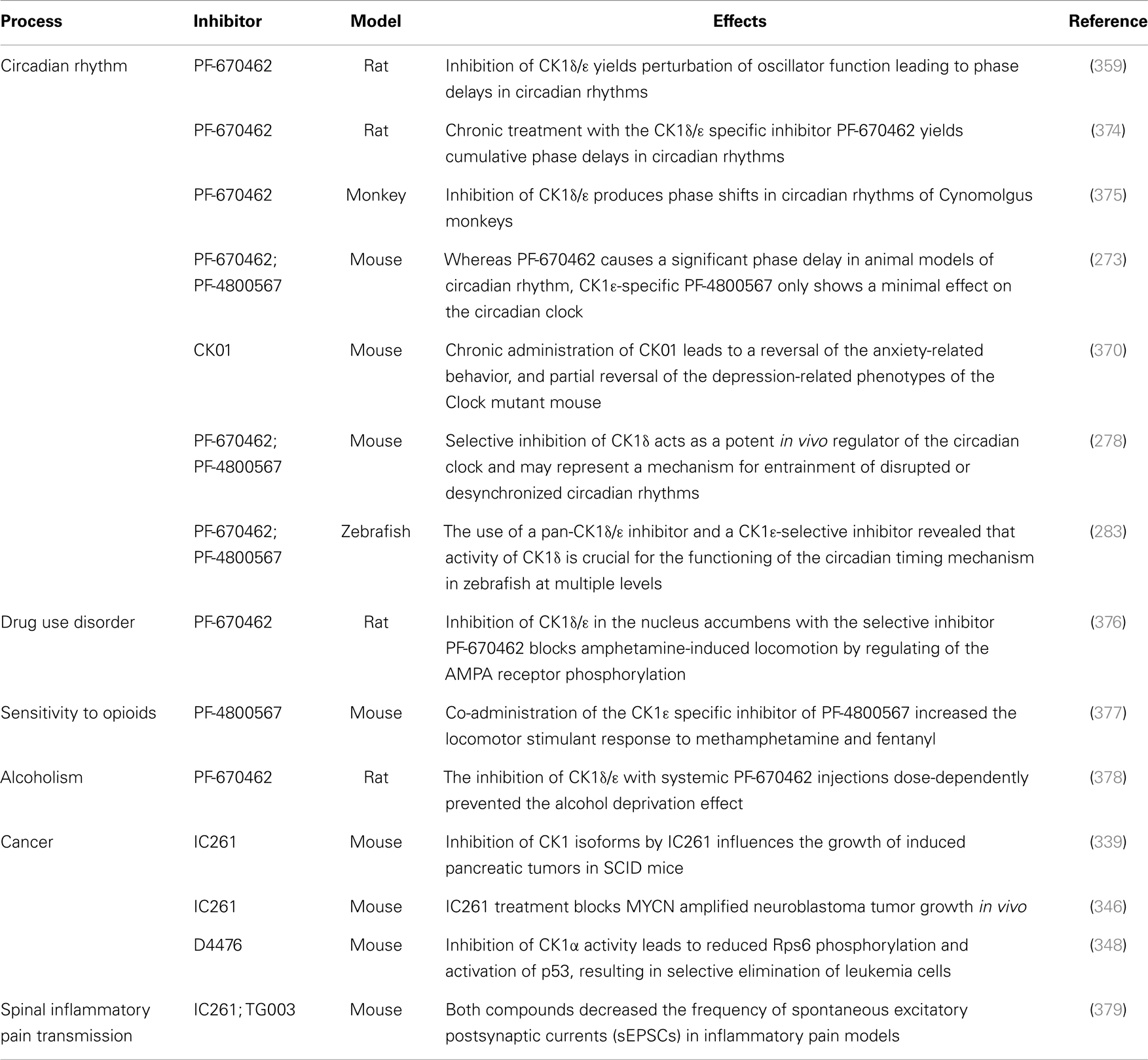
Two very potent and selective inhibitors for CK1δ and ε have been developed by Pfizer Global Research and Development: while PF-670462 possesses only poor isoform selectivity compound PF-4800567 shows a 22-fold stronger inhibition of CK1ε than CK1δ (273, 359). Furthermore, PF-4800567 demonstrated in vivo potency by altering the circadian clock in cycling Rat1 fibroblasts and in a mouse model for circadian rhythm (273). Recently, the use of PF-670462 (and the similar compound CK01) proofed to be beneficial in the treatments of bipolar disorder (370), addictive behavior (378), and in perturbed circadian behavior (278), respectively.
By using structure-based virtual screening Cozza and co-workers identified two amino-anthraquinone analogs as CK1δ-specific inhibitors (362). Furthermore, several roscovitine-derivatives, among them (R)-DRF053, have been shown to inhibit both CK1 and CDK family members (360). In 2009, imidazole- (compounds 17 and 18) and isoxazole-derivatives have been found to be highly potent inhibitors for CK1δ and ε (31). Furthermore, a 2-phenylamino-6-cyano-1H-benzimidazole derivate (compound 1h) was identified as CK1γ-specific inhibitor with excellent selectivity, cellular potency, and acceptable pharmacokinetic properties (368). A new lead compound (a N6-phenyl-1H-pyrazolo[3,4-d]pyrimidine-3,6-diamine derivative), which inhibits CK1 with an IC50 value of 0.078 μM was discovered by Yang and colleagues (369). By using a pyrazolo-pyridine analog as CK1/Chk1 dual-specific inhibitor the p53 pathway could be stabilized and reactivated (MRT00033659) (371). Benzimidazole-based CK1-specific inhibitors were reported by several recent studies (Table 3) [SR-3029 and SR-2890 (366), Bischof-5 and Bischof-6 (367), and Hua-1h (368)]. Furthermore, N-(benzothiazolyl)-2-phenyl-acetamides have been characterized as inhibitors for CK1δ-mediated phosphorylation of TDP-43 and may offer new therapeutic possibilities for the treatment of amyotrophic lateral sclerosis (ALS) (373). Quite recently, also the Clk-specific inhibitor TG003 was used to inhibit CK1 isoforms in a mouse model for mechanical allodynia and thermal hyperalgesia (Table 4) (379).
The potential of CK1-specific inhibitors for the treatment of neuro-degenerative diseases, like AD and Parkinson’s disease, have been recently reviewed in detail by Perez and colleagues (382). The involvement of CK1 isoforms in the pathogenesis of AD is illustrated in Figure 9.
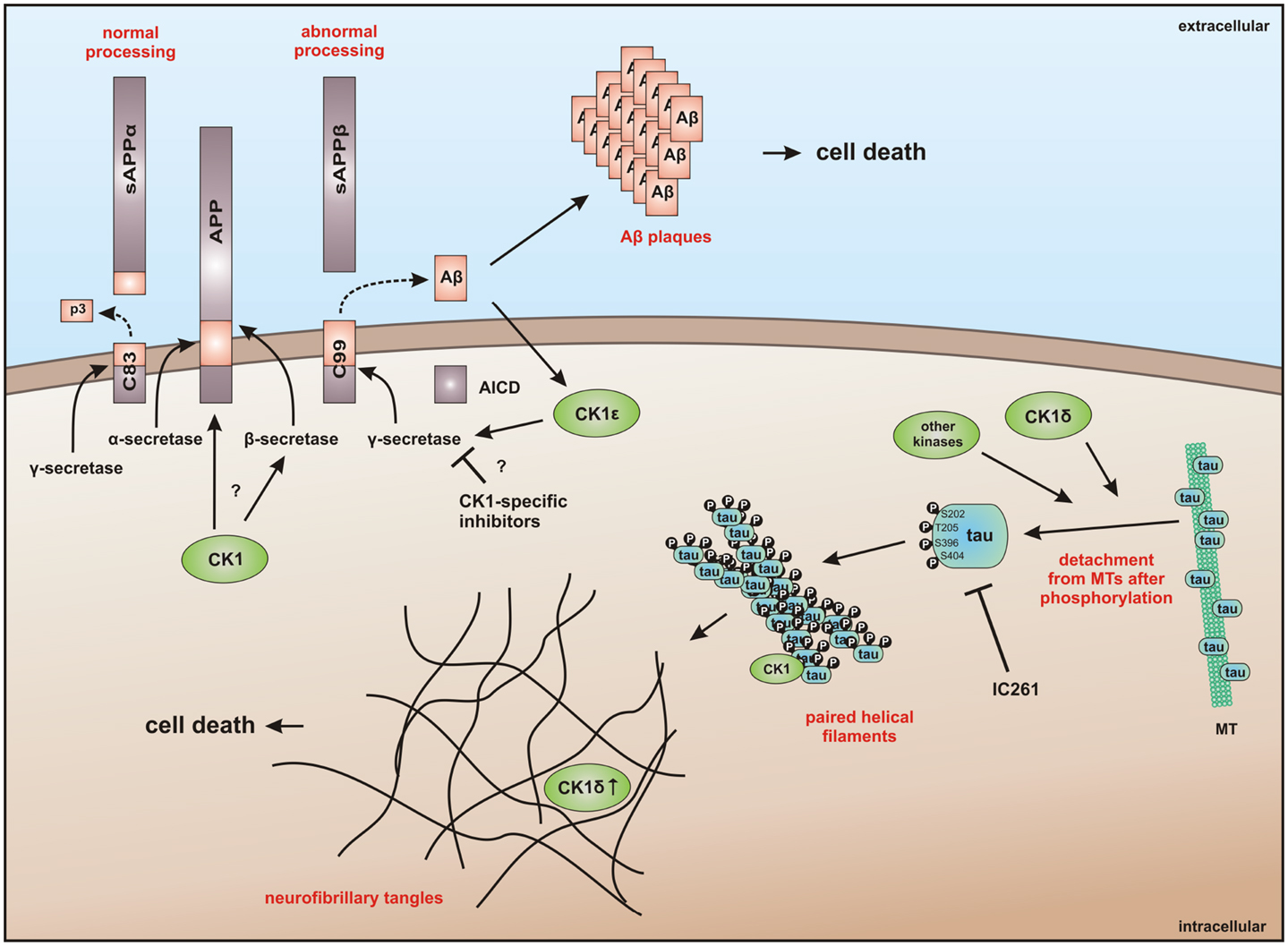
Figure 9. CK1 in neuro-degenerative diseases. It has generally been accepted that overexpression of CK1 plays an important role in neuro-degenerative diseases, especially in tauopathies, such as Alzheimer’s disease (AD). CK1δ is known to be up-regulated up to 30-fold on mRNA level in hippocampal regions of Alzheimer‘s disease (AD) brains (383). CK1δ plays a critical role in formation of neurofibrillary tangles through phosphorylation of tau at amino acids Ser-202/Thr-205 and Ser-396/Ser-404 (responsible for binding to tubulin) in human embryonic kidney 293 cells, thereby leading to a release of tau from MT and to destablilization of MT. Phosphorylation of these sites could be inhibited by the CK1-specific inhibitor IC261 (227). It is further known, that CK1 is associated to paired helical filaments in AD (384) and to tau-containing neurofibrillary tangles, in AD, Down syndrome, progressive supranuclear palsy, Parkinsonism–dementia complex and pallido-ponto-nigral degeneration (383, 385). The overexpression of constitutively active CK1ε, proposed to be involved in processing of amyloid precursor protein (APP) on γ-secretase level, results in an increase of amyloid-beta (Aβ) production, which is attenuated by use of CK1-specific inhibitors (386). In addition, Höttecke et al. (381) could show that the inhibition of γ-secretase by one of these inhibitors does not depend on CK1δ. An in silico analysis further revealed multiple CK1 consensus phosphorylation sites in the intracellular regions of APP, β-secretase, and γ-secretase subunits. Conversely, Aβ seems to influence CK1 activity (387). sAPPα/β: secreted amyloid precursor protein α/β; AICD: amyloid precursor protein intracellular domain.
As an alternative to small molecule inhibitors lacking appropriate ADME (absorption, distribution, metabolism, and excretion) properties or showing unfavorable side effects synthetic peptides can also be used, which copy naturally occurring motifs that specifically influence the activity of the kinase or its interaction with cellular binding partners (388). Lately, small CK1α-derived peptides were used as Biologic tools to block CK1α binding to MDM2. At least, one peptide was identified to block the CK1α-MDM2 interaction (but not CK1α kinase activity) thus leading to decreased CK1α-MDM2-mediated degradation of p53 (208).
Final Remarks
Summarizing the findings cumulated within many years regarding CK1 and its cellular functions, CK1 isoforms can be seen as central players in the regulation of numerous physiological cellular processes. Respecting this involvement in important cellular signal transduction pathways, it is reasonable that dysregulation of CK1 isoforms has been linked to the incidence of inflammatory and proliferative diseases but also to neuro-degenerative disorders. A summary of CK1-associated functions in neuro-degenerative diseases can be found in Figure 9 and its associated figure legend. If potent CK1 (isoform)-specific inhibitors were available new therapeutic possibilities for personalized medicine could be provided. However, the development of isoform-selective compounds available for in vivo application still remains challenging and inhibitor development should include not only conventional small molecule design, but also novel peptide inhibitor approaches.
Authors Contribution
All Authors (Uwe Knippschild, Marc Krüger, Julia Richter, Pengfei Xu, Balbina García-Reyes, Christian Peifer, Jakob Halekotte, Vasiliy Bakulev, and Joachim Bischof) were involved in writing passages of the present review article and participated in final approval and revision. Figures and tables were created by Uwe Knippschild, Marc Krüger, Julia Richter, Pengfei Xu, Balbina García-Reyes, Christian Peifer, Jakob Halekotte, Vasiliy Bakulev, and Joachim Bischof.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Annette Blatz for excellent technical support and PD Dr. Klaus Kramer and PD Dr. Anna Maria Wolf for helpful discussions and critically reading the manuscript. Work in the lab of Uwe Knippschild is funded by the Deutsche Forschungsgemeinschaft (DFG) (KN356/6-1).
References
1. Bingham KD, Shaw DA. The metabolism of testosterone by human male scalp skin. J Endocrinol (1973) 57(1):111–21. doi: 10.1677/joe.0.0570111
2. Agostinis P, Pinna LA, Meggio F, Marin O, Goris J, Vandenheede JR, et al. A synthetic peptide substrate specific for casein kinase I. FEBS Lett (1989) 259(1):75–8. doi:10.1016/0014-5793(89)81498-X
3. Flotow H, Graves PR, Wang AQ, Fiol CJ, Roeske RW, Roach PJ. Phosphate groups as substrate determinants for casein kinase I action. J Biol Chem (1990) 265(24):14264–9.
4. Flotow H, Roach PJ. Role of acidic residues as substrate determinants for casein kinase I. J Biol Chem (1991) 266(6):3724–7.
5. Graves PR, Haas DW, Hagedorn CH, DePaoli-Roach AA, Roach PJ. Molecular cloning, expression, and characterization of a 49-kilodalton casein kinase I isoform from rat testis. J Biol Chem (1993) 268(9):6394–401.
6. Meggio F, Perich JW, Marin O, Pinna LA. The comparative efficiencies of the Ser(P)-, Thr(P)- and Tyr(P)-residues as specificity determinants for casein kinase-1. Biochem Biophys Res Commun (1992) 182(3):1460–5. doi:10.1016/0006-291X(92)91898-Z
7. Meggio F, Perich JW, Reynolds EC, Pinna LA. A synthetic beta-casein phosphopeptide and analogues as model substrates for casein kinase-1, a ubiquitous, phosphate directed protein kinase. FEBS Lett (1991) 283(2):303–6. doi:10.1016/0014-5793(91)80614-9
8. Fish KJ, Cegielska A, Getman ME, Landes GM, Virshup DM. Isolation and characterization of human casein kinase I epsilon (CKI), a novel member of the CKI gene family. J Biol Chem (1995) 270(25):14875–83. doi:10.1074/jbc.270.25.14875
9. Gross SD, Anderson RA. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell Signal (1998) 10(10):699–711. doi:10.1016/S0898-6568(98)00042-4
10. Hathaway GM, Traugh JA. Cyclic nucleotide-independent protein kinases from rabbit reticulocytes. Purification of casein kinases. J Biol Chem (1979) 254(3):762–8.
11. Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science (2002) 298(5600):1912–34. doi:10.1126/science.1075762
12. Ospina B, Fernandez-Renart M. Characterization of three casein kinases type I from Dictyostelium discoideum. Biochim Biophys Acta (1990) 1052(3):483–8. doi:10.1016/0167-4889(90)90159-B
13. Rowles J, Slaughter C, Moomaw C, Hsu J, Cobb MH. Purification of casein kinase I and isolation of cDNAs encoding multiple casein kinase I-like enzymes. Proc Natl Acad Sci U S A (1991) 88(21):9548–52. doi:10.1073/pnas.88.21.9548
14. Tapia C, Featherstone T, Gomez C, Taillon-Miller P, Allende CC, Allende JE. Cloning and chromosomal localization of the gene coding for human protein kinase CK1. FEBS Lett (1994) 349(2):307–12. doi:10.1016/0014-5793(94)00679-2
15. Walczak CE, Anderson RA, Nelson DL. Identification of a family of casein kinases in Paramecium: biochemical characterization and cellular localization. Biochem J (1993) 296(Pt 3):729–35.
16. Zhai L, Graves PR, Robinson LC, Italiano M, Culbertson MR, Rowles J, et al. Casein kinase I gamma subfamily. Molecular cloning, expression, and characterization of three mammalian isoforms and complementation of defects in the Saccharomyces cerevisiae YCK genes. J Biol Chem (1995) 270(21):12717–24.
17. DeMaggio AJ, Lindberg RA, Hunter T, Hoekstra MF. The budding yeast HRR25 gene product is a casein kinase I isoform. Proc Natl Acad Sci U S A (1992) 89(15):7008–12. doi:10.1073/pnas.89.15.7008
18. Dhillon N, Hoekstra MF. Characterization of two protein kinases from Schizosaccharomyces pombe involved in the regulation of DNA repair. EMBO J (1994) 13(12):2777–88.
19. Kearney PH, Ebert M, Kuret J. Molecular cloning and sequence analysis of two novel fission yeast casein kinase-1 isoforms. Biochem Biophys Res Commun (1994) 203(1):231–6. doi:10.1006/bbrc.1994.2172
20. Robinson LC, Hubbard EJ, Graves PR, DePaoli-Roach AA, Roach PJ, Kung C, et al. Yeast casein kinase I homologues: an essential gene pair. Proc Natl Acad Sci U S A (1992) 89(1):28–32. doi:10.1073/pnas.89.1.28
21. Wang PC, Vancura A, Mitcheson TG, Kuret J. Two genes in Saccharomyces cerevisiae encode a membrane-bound form of casein kinase-1. Mol Biol Cell (1992) 3(3):275–86. doi:10.1091/mbc.3.3.275
22. Wang Y, Liu TB, Patel S, Jiang L, Xue C. The casein kinase I protein Cck1 regulates multiple signaling pathways and is essential for cell integrity and fungal virulence in Cryptococcus neoformans. Eukaryot Cell (2011) 10(11):1455–64. doi:10.1128/EC.05207-11
23. van Ooijen G, Martin SF, Barrios-Llerena ME, Hindle M, Le Bihan T, O’Neill JS, et al. Functional analysis of the rodent CK1tau mutation in the circadian clock of a marine unicellular alga. BMC Cell Biol (2013) 14:46. doi:10.1186/1471-2121-14-46
24. Xu RM, Carmel G, Sweet RM, Kuret J, Cheng X. Crystal structure of casein kinase-1, a phosphate-directed protein kinase. EMBO J (1995) 14(5):1015–23.
25. Longenecker KL, Roach PJ, Hurley TD. Three-dimensional structure of mammalian casein kinase I: molecular basis for phosphate recognition. J Mol Biol (1996) 257(3):618–31. doi:10.1006/jmbi.1996.0189
26. Longenecker KL, Roach PJ, Hurley TD. Crystallographic studies of casein kinase I delta toward a structural understanding of auto-inhibition. Acta Crystallogr D Biol Crystallogr (1998) 54(Pt 3):473–5. doi:10.1107/S0907444997011724
27. Taylor SS, Knighton DR, Zheng J, Sowadski JM, Gibbs CS, Zoller MJ. A template for the protein kinase family. Trends Biochem Sci (1993) 18(3):84–9. doi:10.1016/0968-0004(93)80001-R
28. Carmel G, Leichus B, Cheng X, Patterson SD, Mirza U, Chait BT, et al. Expression, purification, crystallization, and preliminary x-ray analysis of casein kinase-1 from Schizosaccharomyces pombe. J Biol Chem (1994) 269(10):7304–9.
30. Hantschel O, Superti-Furga G. Regulation of the c-Abl and Bcr-Abl tyrosine kinases. Nat Rev Mol Cell Biol (2004) 5(1):33–44. doi:10.1038/nrm1280
31. Peifer C, Abadleh M, Bischof J, Hauser D, Schattel V, Hirner H, et al. 3,4-Diaryl-isoxazoles and -imidazoles as potent dual inhibitors of p38alpha mitogen activated protein kinase and casein kinase 1 delta. J Med Chem (2009) 52(23):7618–30. doi:10.1021/jm9005127
32. Behrend L, Stoter M, Kurth M, Rutter G, Heukeshoven J, Deppert W, et al. Interaction of casein kinase 1 delta (CK1delta) with post-Golgi structures, microtubules and the spindle apparatus. Eur J Cell Biol (2000) 79(4):240–51. doi:10.1078/S0171-9335(04)70027-8
33. Roof DM, Meluh PB, Rose MD. Kinesin-related proteins required for assembly of the mitotic spindle. J Cell Biol (1992) 118(1):95–108. doi:10.1083/jcb.118.1.95
34. Tuazon PT, Traugh JA. Casein kinase I and II – multipotential serine protein kinases: structure, function, and regulation. Adv Second Messenger Phosphoprotein Res (1991) 23:123–64.
35. Fu Z, Chakraborti T, Morse S, Bennett GS, Shaw G. Four casein kinase I isoforms are differentially partitioned between nucleus and cytoplasm. Exp Cell Res (2001) 269(2):275–86. doi:10.1006/excr.2001.5324
36. Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, et al. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res (2010) 38(Web Server issue):W695–9. doi:10.1093/nar/gkq313
37. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics (2007) 23(21):2947–8. doi:10.1093/bioinformatics/btm404
38. Bischof J, Randoll SJ, Sussner N, Henne-Bruns D, Pinna LA, Knippschild U. CK1delta kinase activity is modulated by Chk1-mediated phosphorylation. PLoS One (2013) 8(7):e68803. doi:10.1371/journal.pone.0068803
39. Giamas G, Hirner H, Shoshiashvili L, Grothey A, Gessert S, Kuhl M, et al. Phosphorylation of CK1delta: identification of Ser370 as the major phosphorylation site targeted by PKA in vitro and in vivo. Biochem J (2007) 406(3):389–98. doi:10.1042/BJ20070091
40. Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, et al. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol Cell (2008) 31(3):438–48. doi:10.1016/j.molcel.2008.07.007
41. Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, et al. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A (2008) 105(31):10762–7. doi:10.1073/pnas.0805139105
42. Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res (2012) 40(Database issue):D261–70. doi:10.1093/nar/gkr1122
43. Olsen JV, Macek B. High accuracy mass spectrometry in large-scale analysis of protein phosphorylation. Methods Mol Biol (2009) 492:131–42. doi:10.1007/978-1-59745-493-3_7
44. Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, Keri G, et al. Large-scale proteomics analysis of the human kinome. Mol Cell Proteomics (2009) 8(7):1751–64. doi:10.1074/mcp.M800588-MCP200
45. Phanstiel DH, Brumbaugh J, Wenger CD, Tian S, Probasco MD, Bailey DJ, et al. Proteomic and phosphoproteomic comparison of human ES and iPS cells. Nat Methods (2011) 8(10):821–7. doi:10.1038/nmeth.1699
46. Shiromizu T, Adachi J, Watanabe S, Murakami T, Kuga T, Muraoka S, et al. Identification of Missing Proteins in the neXtProt Database and Unregistered Phosphopeptides in the PhosphoSitePlus Database As Part of the Chromosome-Centric Human Proteome Project. J Proteome Res (2013) 12(6):2414–21. doi:10.1021/pr300825v
47. Weber C, Schreiber TB, Daub H. Dual phosphoproteomics and chemical proteomics analysis of erlotinib and gefitinib interference in acute myeloid leukemia cells. J Proteomics (2012) 75(4):1343–56. doi:10.1016/j.jprot.2011.11.004
48. Wissing J, Jansch L, Nimtz M, Dieterich G, Hornberger R, Keri G, et al. Proteomics analysis of protein kinases by target class-selective prefractionation and tandem mass spectrometry. Mol Cell Proteomics (2007) 6(3):537–47. doi:10.1074/mcp.T600062-MCP200
49. Mayya V, Lundgren DH, Hwang SI, Rezaul K, Wu L, Eng JK, et al. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci Signal (2009) 2(84):ra46. doi:10.1126/scisignal.2000007
50. Van Hoof D, Munoz J, Braam SR, Pinkse MW, Linding R, Heck AJ, et al. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell (2009) 5(2):214–26. doi:10.1016/j.stem.2009.05.021
51. Christensen GL, Kelstrup CD, Lyngso C, Sarwar U, Bogebo R, Sheikh SP, et al. Quantitative phosphoproteomics dissection of seven-transmembrane receptor signaling using full and biased agonists. Mol Cell Proteomics (2010) 9(7):1540–53. doi:10.1074/mcp.M900550-MCP200
52. Schreiber TB, Mausbacher N, Keri G, Cox J, Daub H. An integrated phosphoproteomics work flow reveals extensive network regulation in early lysophosphatidic acid signaling. Mol Cell Proteomics (2010) 9(6):1047–62. doi:10.1074/mcp.M900486-MCP200
53. Demirkan G, Yu K, Boylan JM, Salomon AR, Gruppuso PA. Phosphoproteomic profiling of in vivo signaling in liver by the mammalian target of rapamycin complex 1 (mTORC1). PLoS One (2011) 6(6):e21729. doi:10.1371/journal.pone.0021729
54. Marin O, Bustos VH, Cesaro L, Meggio F, Pagano MA, Antonelli M, et al. A noncanonical sequence phosphorylated by casein kinase 1 in beta-catenin may play a role in casein kinase 1 targeting of important signaling proteins. Proc Natl Acad Sci U S A (2003) 100(18):10193–200. doi:10.1073/pnas.1733909100
55. Kawakami F, Suzuki K, Ohtsuki K. A novel consensus phosphorylation motif in sulfatide- and cholesterol-3-sulfate-binding protein substrates for CK1 in vitro. Biol Pharm Bull (2008) 31(2):193–200. doi:10.1248/bpb.31.193
56. Singh TJ, Akatsuka A, Huang KP, Sharma RK, Tam SW, Wang JH. A multifunctional cyclic nucleotide- and Ca2+-independent protein kinase from rabbit skeletal muscle. Biochem Biophys Res Commun (1982) 107(2):676–83. doi:10.1016/0006-291X(82)91544-3
57. Lu PW, Soong CJ, Tao M. Phosphorylation of ankyrin decreases its affinity for spectrin tetramer. J Biol Chem (1985) 260(28):14958–64.
58. Eder PS, Soong CJ, Tao M. Phosphorylation reduces the affinity of protein 4.1 for spectrin. Biochemistry (1986) 25(7):1764–70. doi:10.1021/bi00355a047
59. Nakajo S, Nakaya K, Nakamura Y. Phosphorylation of actin-binding proteins by casein kinases 1 and 2. Biochem Int (1987) 15(2):321–7.
60. Floyd CC, Grant P, Gallant PE, Pant HC. Principal neurofilament-associated protein kinase in squid axoplasm is related to casein kinase I. J Biol Chem (1991) 266(8):4987–94.
61. Link WT, Dosemeci A, Floyd CC, Pant HC. Bovine neurofilament-enriched preparations contain kinase activity similar to casein kinase I – neurofilament phosphorylation by casein kinase I (CKI). Neurosci Lett (1993) 151(1):89–93. doi:10.1016/0304-3940(93)90053-N
62. Yang P, Sale WS. Casein kinase I is anchored on axonemal doublet microtubules and regulates flagellar dynein phosphorylation and activity. J Biol Chem (2000) 275(25):18905–12. doi:10.1074/jbc.M002134200
63. Wolff S, Xiao Z, Wittau M, Sussner N, Stoter M, Knippschild U. Interaction of casein kinase 1 delta (CK1delta) with the light chain LC2 of microtubule associated protein 1A (MAP1A). Biochim Biophys Acta (2005) 1745(2):196–206. doi:10.1016/j.bbamcr.2005.05.004
64. Singh TJ, Grundke-Iqbal I, Iqbal K. Phosphorylation of tau protein by casein kinase-1 converts it to an abnormal Alzheimer-like state. J Neurochem (1995) 64(3):1420–3. doi:10.1046/j.1471-4159.1995.64031420.x
65. Gao ZH, Metherall J, Virshup DM. Identification of casein kinase I substrates by in vitro expression cloning screening. Biochem Biophys Res Commun (2000) 268(2):562–6. doi:10.1006/bbrc.2000.2168
66. Dubois T, Howell S, Zemlickova E, Aitken A. Identification of casein kinase I alpha interacting protein partners. FEBS Lett (2002) 517(1–3):167–71. doi:10.1016/S0014-5793(02)02614-5
67. Dubois T, Kerai P, Learmonth M, Cronshaw A, Aitken A. Identification of syntaxin-1A sites of phosphorylation by casein kinase I and casein kinase II. Eur J Biochem (2002) 269(3):909–14. doi:10.1046/j.0014-2956.2001.02725.x
68. Mackie K, Sorkin BC, Nairn AC, Greengard P, Edelman GM, Cunningham BA. Identification of two protein kinases that phosphorylate the neural cell-adhesion molecule, N-CAM. J Neurosci (1989) 9(6):1883–96.
69. Dupre-Crochet S, Figueroa A, Hogan C, Ferber EC, Bialucha CU, Adams J, et al. Casein kinase 1 is a novel negative regulator of E-cadherin-based cell-cell contacts. Mol Cell Biol (2007) 27(10):3804–16. doi:10.1128/MCB.01590-06
70. Tillement V, Lajoie-Mazenc I, Casanova A, Froment C, Penary M, Tovar D, et al. Phosphorylation of RhoB by CK1 impedes actin stress fiber organization and epidermal growth factor receptor stabilization. Exp Cell Res (2008) 314(15):2811–21. doi:10.1016/j.yexcr.2008.06.011
71. Mennella V, Tan DY, Buster DW, Asenjo AB, Rath U, Ma A, et al. Motor domain phosphorylation and regulation of the Drosophila kinesin 13, KLP10A. J Cell Biol (2009) 186(4):481–90. doi:10.1083/jcb.200902113
72. Huflejt ME, Turck CW, Lindstedt R, Barondes SH, Leffler H. L-29, a soluble lactose-binding lectin, is phosphorylated on serine 6 and serine 12 in vivo and by casein kinase I. J Biol Chem (1993) 268(35):26712–8.
73. Szabo P, Dam TK, Smetana K Jr, Dvorankova B, Kubler D, Brewer CF, et al. Phosphorylated human lectin galectin-3: analysis of ligand binding by histochemical monitoring of normal/malignant squamous epithelia and by isothermal titration calorimetry. Anat Histol Embryol (2009) 38(1):68–75. doi:10.1111/j.1439-0264.2008.00899.x
74. Zyss D, Ebrahimi H, Gergely F. Casein kinase I delta controls centrosome positioning during T cell activation. J Cell Biol (2011) 195(5):781–97. doi:10.1083/jcb.201106025
75. Johnson AE, Chen JS, Gould KL. CK1 is required for a mitotic checkpoint that delays cytokinesis. Curr Biol (2013) 23(19):1920–6. doi:10.1016/j.cub.2013.07.077
76. Cooper CD, Lampe PD. Casein kinase 1 regulates connexin-43 gap junction assembly. J Biol Chem (2002) 277(47):44962–8. doi:10.1074/jbc.M209427200
77. Zhong J, Shaik S, Wan L, Tron AE, Wang Z, Sun L, et al. SCFbeta-TRCP targets MTSS1 for ubiquitination-mediated destruction to regulate cancer cell proliferation and migration. Oncotarget (2013) 4(12):2339–53.
78. Muller P, Ruckova E, Halada P, Coates PJ, Hrstka R, Lane DP, et al. C-terminal phosphorylation of Hsp70 and Hsp90 regulates alternate binding to co-chaperones CHIP and HOP to determine cellular protein folding/degradation balances. Oncogene (2013) 32(25):3101–10. doi:10.1038/onc.2012.314
79. Rapuano M, Rosen OM. Phosphorylation of the insulin receptor by a casein kinase I-like enzyme. J Biol Chem (1991) 266(20):12902–7.
80. Darnay BG, Singh S, Aggarwal BB. The p80 TNF receptor-associated kinase (p80TRAK) associates with residues 354-397 of the p80 cytoplasmic domain: similarity to casein kinase. FEBS Lett (1997) 406(1–2):101–5. doi:10.1016/S0014-5793(97)00251-2
81. Tobin AB, Totty NF, Sterlin AE, Nahorski SR. Stimulus-dependent phosphorylation of G-protein-coupled receptors by casein kinase 1alpha. J Biol Chem (1997) 272(33):20844–9. doi:10.1074/jbc.272.33.20844
82. Hicke L, Zanolari B, Riezman H. Cytoplasmic tail phosphorylation of the alpha-factor receptor is required for its ubiquitination and internalization. J Cell Biol (1998) 141(2):349–58. doi:10.1083/jcb.141.2.349
83. Feng Y, Davis NG. Akr1p and the type I casein kinases act prior to the ubiquitination step of yeast endocytosis: Akr1p is required for kinase localization to the plasma membrane. Mol Cell Biol (2000) 20(14):5350–9. doi:10.1128/MCB.20.14.5350-5359.2000
84. Bioukar EB, Marricco NC, Zuo D, Larose L. Serine phosphorylation of the ligand-activated beta-platelet-derived growth factor receptor by casein kinase I-gamma2 inhibits the receptor’s autophosphorylating activity. J Biol Chem (1999) 274(30):21457–63. doi:10.1074/jbc.274.30.21457
85. Zhao Y, Qin S, Atangan LI, Molina Y, Okawa Y, Arpawong HT, et al. Casein Kinase 1{alpha} Interacts with Retinoid X Receptor and Interferes with Agonist-induced Apoptosis. J Biol Chem (2004) 279(29):30844–9. doi:10.1074/jbc.M404651200
86. Davidson G, Wu W, Shen J, Bilic J, Fenger U, Stannek P, et al. Casein kinase 1 gamma couples Wnt receptor activation to cytoplasmic signal transduction. Nature (2005) 438(7069):867–72. doi:10.1038/nature04170
87. Zeng X, Tamai K, Doble B, Li S, Huang H, Habas R, et al. A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature (2005) 438(7069):873–7. doi:10.1038/nature04185
88. Liu J, Carvalho LP, Bhattacharya S, Carbone CJ, Kumar KG, Leu NA, et al. Mammalian casein kinase 1alpha and its leishmanial ortholog regulate stability of IFNAR1 and type I interferon signaling. Mol Cell Biol (2009) 29(24):6401–12. doi:10.1128/MCB.00478-09
89. Giamas G, Castellano L, Feng Q, Knippschild U, Jacob J, Thomas RS, et al. CK1delta modulates the transcriptional activity of ERalpha via AIB1 in an estrogen-dependent manner and regulates ERalpha-AIB1 interactions. Nucleic Acids Res (2009) 37(9):3110–23. doi:10.1093/nar/gkp136
90. Papoff G, Trivieri N, Crielesi R, Ruberti F, Marsilio S, Ruberti G. FADD-calmodulin interaction: a novel player in cell cycle regulation. Biochim Biophys Acta (2010) 1803(8):898–911. doi:10.1016/j.bbamcr.2010.04.006
91. Kani S, Oishi I, Yamamoto H, Yoda A, Suzuki H, Nomachi A, et al. The receptor tyrosine kinase Ror2 associates with and is activated by casein kinase Iepsilon. J Biol Chem (2004) 279(48):50102–9. doi:10.1074/jbc.M409039200
92. Soong CJ, Lu PW, Tao M. Analysis of band 3 cytoplasmic domain phosphorylation and association with ankyrin. Arch Biochem Biophys (1987) 254(2):509–17. doi:10.1016/0003-9861(87)90131-7
93. Marchal C, Dupre S, Urban-Grimal D. Casein kinase I controls a late step in the endocytic trafficking of yeast uracil permease. J Cell Sci (2002) 115(Pt 1):217–26.
94. Gerbeth C, Schmidt O, Rao S, Harbauer AB, Mikropoulou D, Opalinska M, et al. Glucose-induced regulation of protein import receptor Tom22 by cytosolic and mitochondria-bound kinases. Cell Metab (2013) 18(4):578–87. doi:10.1016/j.cmet.2013.09.006
95. Yan W, Spruce L, Rosenblatt MM, Kleyman TR, Rubenstein RC. Intracellular trafficking of a polymorphism in the COOH terminus of the alpha-subunit of the human epithelial sodium channel is modulated by casein kinase 1. Am J Physiol Renal Physiol (2007) 293(3):F868–76. doi:10.1152/ajprenal.00194.2007
96. Christmann JL, Dahmus ME. Phosphorylation of rat ascites tumor non-histone chromatin proteins. Differential phosphorylation by two cyclic nucleotide-independent protein kinases and comparison to in vivo phosphorylation. J Biol Chem (1981) 256(7):3326–31.
97. Dahmus ME. Purification and properties of calf thymus casein kinases I and II. J Biol Chem (1981) 256(7):3319–25.
98. Grozav AG, Chikamori K, Kozuki T, Grabowski DR, Bukowski RM, Willard B, et al. Casein kinase I delta/epsilon phosphorylates topoisomerase IIalpha at serine-1106 and modulates DNA cleavage activity. Nucleic Acids Res (2009) 37(2):382–92. doi:10.1093/nar/gkn934
99. Gonzales ML, Mellman DL, Anderson RA. CKIalpha is associated with and phosphorylates star-PAP and is also required for expression of select star-PAP target messenger RNAs. J Biol Chem (2008) 283(18):12665–73. doi:10.1074/jbc.M800656200
100. Ishiguro T, Tanaka K, Sakuno T, Watanabe Y. Shugoshin-PP2A counteracts casein-kinase-1-dependent cleavage of Rec8 by separase. Nat Cell Biol (2010) 12(5):500–6. doi:10.1038/ncb2052
101. Sugiyama Y, Hatano N, Sueyoshi N, Suetake I, Tajima S, Kinoshita E, et al. The DNA-binding activity of mouse DNA methyltransferase 1 is regulated by phosphorylation with casein kinase 1 delta/epsilon. Biochem J (2010) 427(3):489–97. doi:10.1042/BJ20091856
102. Choksi DK, Roy B, Chatterjee S, Yusuff T, Bakhoum MF, Sengupta U, et al. TDP-43 Phosphorylation by casein kinase I{varepsilon} promotes oligomerization and enhances toxicity in vivo. Hum Mol Genet (2014) 23(4):1025–35. doi:10.1093/hmg/ddt498
103. Cruciat CM, Dolde C, de Groot RE, Ohkawara B, Reinhard C, Korswagen HC, et al. RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in Wnt/beta-catenin signaling. Science (2013) 339(6126):1436–41. doi:10.1126/science.1231499
104. Chen H, Ma H, Inuzuka H, Diao J, Lan F, Shi YG, et al. DNA damage regulates UHRF1 stability via the SCF(beta-TrCP) E3 ligase. Mol Cell Biol (2013) 33(6):1139–48. doi:10.1128/MCB.01191-12
105. Wojda I, Cytrynska M, Frajnt M, Jakubowicz T. Phosphorylation of yeast ribosomal proteins by CKI and CKII in the presence of heparin. Acta Biochim Pol (1999) 46(1):211–5.
106. Hutchinson JA, Shanware NP, Chang H, Tibbetts RS. Regulation of ribosomal protein S6 phosphorylation by casein kinase 1 and protein phosphatase 1. J Biol Chem (2011) 286(10):8688–96. doi:10.1074/jbc.M110.141754
107. Zemp I, Wandrey F, Rao S, Ashiono C, Wyler E, Montellese C, et al. CK1δ and CK1ε are components of human 40S subunit precursors required for cytoplasmic 40S maturation. J Cell Sci (2014) 127(Pt 6):1242–53. doi:10.1242/jcs.138719
108. Milne DM, Palmer RH, Campbell DG, Meek DW. Phosphorylation of the p53 tumour-suppressor protein at three N-terminal sites by a novel casein kinase I-like enzyme. Oncogene (1992) 7(7):1361–9.
109. de Groot RP, den Hertog J, Vandenheede JR, Goris J, Sassone-Corsi P. Multiple and cooperative phosphorylation events regulate the CREM activator function. EMBO J (1993) 12(10):3903–11.
110. Ho Y, Mason S, Kobayashi R, Hoekstra M, Andrews B. Role of the casein kinase I isoform, Hrr25, and the cell cycle-regulatory transcription factor, SBF, in the transcriptional response to DNA damage in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A (1997) 94(2):581–6. doi:10.1073/pnas.94.2.581
111. Zhu J, Shibasaki F, Price R, Guillemot JC, Yano T, Dotsch V, et al. Intramolecular masking of nuclear import signal on NF-AT4 by casein kinase I and MEKK1. Cell (1998) 93(5):851–61. doi:10.1016/S0092-8674(00)81445-2
112. Gross SD, Loijens JC, Anderson RA. The casein kinase Ialpha isoform is both physically positioned and functionally competent to regulate multiple events of mRNA metabolism. J Cell Sci (1999) 112(Pt 16):2647–56.
113. Lee E, Salic A, Kirschner MW. Physiological regulation of [beta]-catenin stability by Tcf3 and CK1 epsilon. J Cell Biol (2001) 154(5):983–93. doi:10.1083/jcb.200102074
114. Eide EJ, Vielhaber EL, Hinz WA, Virshup DM. The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iepsilon. J Biol Chem (2002) 277(19):17248–54. doi:10.1074/jbc.M111466200
115. Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, et al. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell (2002) 108(6):837–47. doi:10.1016/S0092-8674(02)00685-2
116. Sakanaka C. Phosphorylation and regulation of beta-catenin by casein kinase I epsilon. J Biochem (2002) 132(5):697–703. doi:10.1093/oxfordjournals.jbchem.a003276
117. Yanagawa S, Matsuda Y, Lee JS, Matsubayashi H, Sese S, Kadowaki T, et al. Casein kinase I phosphorylates the Armadillo protein and induces its degradation in Drosophila. EMBO J (2002) 21(7):1733–42. doi:10.1093/emboj/21.7.1733
118. Waddell DS, Liberati NT, Guo X, Frederick JP, Wang XF. Casein kinase Iepsilon plays a functional role in the transforming growth factor-beta signaling pathway. J Biol Chem (2004) 279(28):29236–46. doi:10.1074/jbc.M400880200
119. Xu S, Wong CC, Tong EH, Chung SS, Yates JR III, Yin Y, et al. Phosphorylation by casein kinase 1 regulates tonicity-induced osmotic response element-binding protein/tonicity enhancer-binding protein nucleocytoplasmic trafficking. J Biol Chem (2008) 283(25):17624–34. doi:10.1074/jbc.M800281200
120. Jia J, Zhang L, Zhang Q, Tong C, Wang B, Hou F, et al. Phosphorylation by double-time/CKIepsilon and CKIalpha targets cubitus interruptus for Slimb/beta-TRCP-mediated proteolytic processing. Dev Cell (2005) 9(6):819–30. doi:10.1016/j.devcel.2005.10.006
121. Regad T, Roth M, Bredenkamp N, Illing N, Papalopulu N. The neural progenitor-specifying activity of FoxG1 is antagonistically regulated by CKI and FGF. Nat Cell Biol (2007) 9(5):531–40. doi:10.1038/ncb1573
122. Xu Y, Lee SH, Kim HS, Kim NH, Piao S, Park SH, et al. Role of CK1 in GSK3beta-mediated phosphorylation and degradation of snail. Oncogene (2010) 29(21):3124–33. doi:10.1038/onc.2010.77
123. Liu CY, Zha ZY, Zhou X, Zhang H, Huang W, Zhao D, et al. The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCF{beta}-TrCP E3 ligase. J Biol Chem (2010) 285(48):37159–69. doi:10.1074/jbc.M110.152942
124. Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Dev (2010) 24(1):72–85. doi:10.1101/gad.1843810
125. Li S, Chen XW, Yu L, Saltiel AR, Lin JD. Circadian metabolic regulation through crosstalk between casein kinase 1 delta and transcriptional coactivator PGC-1alpha. Mol Endocrinol (2011) 25(12):2084–93. doi:10.1210/me.2011-1227
126. Galletti M, Riccardo S, Parisi F, Lora C, Saqcena MK, Rivas L, et al. Identification of domains responsible for ubiquitin-dependent degradation of dMyc by glycogen synthase kinase 3beta and casein kinase 1 kinases. Mol Cell Biol (2009) 29(12):3424–34. doi:10.1128/MCB.01535-08
127. Shanware NP, Trinh AT, Williams LM, Tibbetts RS. Coregulated ataxia telangiectasia-mutated and casein kinase sites modulate cAMP-response element-binding protein-coactivator interactions in response to DNA damage. J Biol Chem (2007) 282(9):6283–91. doi:10.1074/jbc.M610674200
128. Brookheart RT, Lee CY, Espenshade PJ. Casein kinase 1 regulates sterol regulatory element-binding protein (SREBP) to control sterol homeostasis. J Biol Chem (2014) 289(5):2725–35. doi:10.1074/jbc.M113.511899
129. Wang Y, Hu L, Tong X, Ye X. Casein kinase 1 gamma 1 inhibits the RIG-I/TLR signaling pathway through phosphorylating p65 and promoting its degradation. J Immunol (2014) 192(4):1855–61. doi:10.4049/jimmunol.1302552
130. Haas DW, Hagedorn CH. Casein kinase I phosphorylates the 25-kDa mRNA cap-binding protein. Arch Biochem Biophys (1991) 284(1):84–9. doi:10.1016/0003-9861(91)90267-M
131. Biswas A, Mukherjee S, Das S, Shields D, Chow CW, Maitra U. Opposing action of casein kinase 1 and calcineurin in nucleo-cytoplasmic shuttling of mammalian translation initiation factor eIF6. J Biol Chem (2011) 286(4):3129–38. doi:10.1074/jbc.M110.188565
132. Grasser FA, Scheidtmann KH, Tuazon PT, Traugh JA, Walter G. In vitro phosphorylation of SV40 large T antigen. Virology (1988) 165(1):13–22. doi:10.1016/0042-6822(88)90653-8
133. Quintavalle M, Sambucini S, Summa V, Orsatti L, Talamo F, De Francesco R, et al. Hepatitis C virus NS5A is a direct substrate of casein kinase I-alpha, a cellular kinase identified by inhibitor affinity chromatography using specific NS5A hyperphosphorylation inhibitors. J Biol Chem (2007) 282(8):5536–44. doi:10.1074/jbc.M610486200
134. Alvisi G, Marin O, Pari G, Mancini M, Avanzi S, Loregian A, et al. Multiple phosphorylation sites at the C-terminus regulate nuclear import of HCMV DNA polymerase processivity factor ppUL44. Virology (2011) 417(2):259–67. doi:10.1016/j.virol.2011.06.015
135. Makarov VV, Iconnikova AY, Guseinov MA, Vishnichenko VK, Kalinina NO. In vitro phosphorylation of the N-terminal half of hordeivirus movement protein. Biochemistry (Mosc) (2012) 77(9):1072–81. doi:10.1134/S0006297912090155
136. Woodard C, Shamay M, Liao G, Zhu J, Ng AN, Li R, et al. Phosphorylation of the chromatin binding domain of KSHV LANA. PLoS Pathog (2012) 8(10):e1002972. doi:10.1371/journal.ppat.1002972
137. Bhattacharya D, Ansari IH, Striker R. The flaviviral methyltransferase is a substrate of Casein Kinase 1. Virus Res (2009) 141(1):101–4. doi:10.1016/j.virusres.2009.01.002
138. Sharma P, Sharma M, Amin ND, Albers RW, Pant HC. Regulation of cyclin-dependent kinase 5 catalytic activity by phosphorylation. Proc Natl Acad Sci U S A (1999) 96(20):11156–60. doi:10.1073/pnas.96.20.11156
139. Okano M, Yokoyama T, Miyanaga T, Ohtsuki K. Activation of C-kinase eta through its cholesterol-3-sulfate-dependent phosphorylation by casein kinase I in vitro. Biol Pharm Bull (2004) 27(1):109–12. doi:10.1248/bpb.27.109
140. von Blume J, Knippschild U, Dequiedt F, Giamas G, Beck A, Auer A, et al. Phosphorylation at Ser244 by CK1 determines nuclear localization and substrate targeting of PKD2. EMBO J (2007) 26(22):4619–33. doi:10.1038/sj.emboj.7601891
141. Honaker Y, Piwnica-Worms H. Casein kinase 1 functions as both penultimate and ultimate kinase in regulating Cdc25A destruction. Oncogene (2010) 29(23):3324–34. doi:10.1038/onc.2010.96
142. Pal G, Paraz MT, Kellogg DR. Regulation of Mih1/Cdc25 by protein phosphatase 2A and casein kinase 1. J Cell Biol (2008) 180(5):931–45. doi:10.1083/jcb.200711014
143. Piao S, Lee SJ, Xu Y, Gwak J, Oh S, Park BJ, et al. CK1 epsilon targets Cdc25A for ubiquitin-mediated proteolysis under normal conditions and in response to checkpoint activation. Cell Cycle (2011) 10(3):531–7. doi:10.4161/cc.10.3.14757
144. Li X, Liu J, Gao T. Beta-TrCP-mediated ubiquitination and degradation of PHLPP1 are negatively regulated by Akt. Mol Cell Biol (2009) 29(23):6192–205. doi:10.1128/MCB.00681-09
145. Agostinis P, Marin O, James P, Hendrix P, Merlevede W, Vandenheede JR, et al. Phosphorylation of the phosphatase modulator subunit (inhibitor-2) by casein kinase-1. Identification of the phosphorylation sites. FEBS Lett (1992) 305(2):121–4. doi:10.1016/0014-5793(92)80877-J
146. Marin O, Meggio F, Sarno S, Andretta M, Pinna LA. Phosphorylation of synthetic fragments of inhibitor-2 of protein phosphatase-1 by casein kinase-1 and -2. Evidence that phosphorylated residues are not strictly required for efficient targeting by casein kinase-1. Eur J Biochem (1994) 223(2):647–53. doi:10.1111/j.1432-1033.1994.tb19037.x
147. Desdouits F, Siciliano JC, Greengard P, Girault JA. Dopamine- and cAMP-regulated phosphoprotein DARPP-32: phosphorylation of Ser-137 by casein kinase I inhibits dephosphorylation of Thr-34 by calcineurin. Proc Natl Acad Sci U S A (1995) 92(7):2682–5. doi:10.1073/pnas.92.7.2682
148. Peters JM, McKay RM, McKay JP, Graff JM. Casein kinase I transduces Wnt signals. Nature (1999) 401(6751):345–50. doi:10.1038/43830
149. Vielhaber E, Eide E, Rivers A, Gao ZH, Virshup DM. Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I epsilon. Mol Cell Biol (2000) 20(13):4888–99. doi:10.1128/MCB.20.13.4888-4899.2000
150. Rubinfeld B, Tice DA, Polakis P. Axin-dependent phosphorylation of the adenomatous polyposis coli protein mediated by casein kinase 1 epsilon. J Biol Chem (2001) 276(42):39037–45. doi:10.1074/jbc.M105148200
151. Desagher S, Osen-Sand A, Montessuit S, Magnenat E, Vilbois F, Hochmann A, et al. Phosphorylation of bid by casein kinases I and II regulates its cleavage by caspase 8. Mol Cell (2001) 8(3):601–11. doi:10.1016/S1097-2765(01)00335-5
152. Zemlickova E, Johannes FJ, Aitken A, Dubois T. Association of CPI-17 with protein kinase C and casein kinase I. Biochem Biophys Res Commun (2004) 316(1):39–47. doi:10.1016/j.bbrc.2004.02.014
153. Garzia L, D’Angelo A, Amoresano A, Knauer SK, Cirulli C, Campanella C, et al. Phosphorylation of nm23-H1 by CKI induces its complex formation with h-prune and promotes cell motility. Oncogene (2008) 27(13):1853–64. doi:10.1038/sj.onc.1210822
154. Clokie S, Falconer H, Mackie S, Dubois T, Aitken A. The interaction between casein kinase Ialpha and 14-3-3 is phosphorylation dependent. FEBS J (2009) 276(23):6971–84. doi:10.1111/j.1742-4658.2009.07405.x
155. Winter M, Milne D, Dias S, Kulikov R, Knippschild U, Blattner C, et al. Protein kinase CK1delta phosphorylates key sites in the acidic domain of murine double-minute clone 2 protein (MDM2) that regulate p53 turnover. Biochemistry (2004) 43(51):16356–64. doi:10.1021/bi0489255
156. Chen L, Li C, Pan Y, Chen J. Regulation of p53-MDMX interaction by casein kinase 1 alpha. Mol Cell Biol (2005) 25(15):6509–20. doi:10.1128/MCB.25.15.6509-6520.2005
157. He Q, Cha J, Lee HC, Yang Y, Liu Y. CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev (2006) 20(18):2552–65. doi:10.1101/gad.1463506
158. Huang G, Chen S, Li S, Cha J, Long C, Li L, et al. Protein kinase A and casein kinases mediate sequential phosphorylation events in the circadian negative feedback loop. Genes Dev (2007) 21(24):3283–95. doi:10.1101/gad.1610207
159. Bidere N, Ngo VN, Lee J, Collins C, Zheng L, Wan F, et al. Casein kinase 1alpha governs antigen-receptor-induced NF-kappaB activation and human lymphoma cell survival. Nature (2009) 458(7234):92–6. doi:10.1038/nature07613
160. Dai C, Xue HW. Rice early flowering 1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J (2010) 29(11):1916–27. doi:10.1038/emboj.2010.75
161. Isoda M, Sako K, Suzuki K, Nishino K, Nakajo N, Ohe M, et al. Dynamic regulation of Emi2 by Emi2-bound Cdk1/Plk1/CK1 and PP2A-B56 in meiotic arrest of Xenopus eggs. Dev Cell (2011) 21(3):506–19. doi:10.1016/j.devcel.2011.06.029
162. Meng Z, Capalbo L, Glover DM, Dunphy WG. Role for casein kinase 1 in the phosphorylation of Claspin on critical residues necessary for the activation of Chk1. Mol Biol Cell (2011) 22(16):2834–47. doi:10.1091/mbc.E11-01-0048
163. Shanware NP, Hutchinson JA, Kim SH, Zhan L, Bowler MJ, Tibbetts RS. Casein kinase 1-dependent phosphorylation of familial advanced sleep phase syndrome-associated residues controls PERIOD 2 stability. J Biol Chem (2011) 286(14):12766–74. doi:10.1074/jbc.M111.224014
164. Stavenuiter F, Gale AJ, Heeb MJ. Phosphorylation of protein S by platelet kinases enhances its activated protein C cofactor activity. FASEB J (2013) 27(7):2918–25. doi:10.1096/fj.12-225961
165. Magliozzi R, Low TY, Weijts BG, Cheng T, Spanjaard E, Mohammed S, et al. Control of epithelial cell migration and invasion by the IKKbeta- and CK1alpha-mediated degradation of RAPGEF2. Dev Cell (2013) 27(5):574–85. doi:10.1016/j.devcel.2013.10.023
166. Yim DG, Ghosh S, Guy GR, Virshup DM. Casein kinase 1 regulates Sprouty2 in FGF-ERK signaling. Oncogene (2014). doi:10.1038/onc.2013.564
167. Tipper JP, Bacon GW, Witters LA. Phosphorylation of acetyl-coenzyme A carboxylase by casein kinase I and casein kinase II. Arch Biochem Biophys (1983) 227(2):386–96. doi:10.1016/0003-9861(83)90468-X
168. Kuret J, Woodgett JR, Cohen P. Multisite phosphorylation of glycogen synthase from rabbit skeletal muscle. Identification of the sites phosphorylated by casein kinase-I. Eur J Biochem (1985) 151(1):39–48. doi:10.1111/j.1432-1033.1985.tb09066.x
169. Poulter L, Ang SG, Gibson BW, Williams DH, Holmes CF, Caudwell FB, et al. Analysis of the in vivo phosphorylation state of rabbit skeletal muscle glycogen synthase by fast-atom-bombardment mass spectrometry. Eur J Biochem (1988) 175(3):497–510. doi:10.1111/j.1432-1033.1988.tb14222.x
170. Omnus DJ, Ljungdahl PO. Rts1-protein phosphatase 2A antagonizes Ptr3-mediated activation of the signaling protease Ssy5 by casein kinase I. Mol Biol Cell (2013) 24(9):1480–92. doi:10.1091/mbc.E13-01-0019
171. Liu J, Wan L, Liu P, Inuzuka H, Wang Z, Wei W. SCFβ-TRCP-mediated degradation of NEDD4 inhibits tumorigenesis through modulating the PTEN/Akt signaling pathway. Oncotarget (2014) 5(4):1026–37.
172. Gross SD, Hoffman DP, Fisette PL, Baas P, Anderson RA. A phosphatidylinositol 4,5-bisphosphate-sensitive casein kinase I alpha associates with synaptic vesicles and phosphorylates a subset of vesicle proteins. J Cell Biol (1995) 130(3):711–24. doi:10.1083/jcb.130.3.711
173. Faundez VV, Kelly RB. The AP-3 complex required for endosomal synaptic vesicle biogenesis is associated with a casein kinase I alpha-like isoform. Mol Biol Cell (2000) 11(8):2591–604. doi:10.1091/mbc.11.8.2591
174. Wolff S, Stoter M, Giamas G, Piesche M, Henne-Bruns D, Banting G, et al. Casein kinase 1 delta (CK1delta) interacts with the SNARE associated protein snapin. FEBS Lett (2006) 580(27):6477–84. doi:10.1016/j.febslet.2006.10.068
175. Tomishige N, Kumagai K, Kusuda J, Nishijima M, Hanada K. Casein kinase I{gamma}2 down-regulates trafficking of ceramide in the synthesis of sphingomyelin. Mol Biol Cell (2009) 20(1):348–57. doi:10.1091/mbc.E08-07-0669
176. Alappat EC, Feig C, Boyerinas B, Volkland J, Samuels M, Murmann AE, et al. Phosphorylation of FADD at serine 194 by CKIalpha regulates its nonapoptotic activities. Mol Cell (2005) 19(3):321–32. doi:10.1016/j.molcel.2005.06.024
177. Wang Y, Sun X, Wu J, Xu BE, Gu C, Wang H, et al. Casein kinase 1 alpha interacts with RIP1 and regulates NF-kappaB activation. Biochemistry (2008) 47(1):441–8. doi:10.1021/bi7010515
178. Walter J, Grunberg J, Schindzielorz A, Haass C. Proteolytic fragments of the Alzheimer’s disease associated presenilins-1 and -2 are phosphorylated in vivo by distinct cellular mechanisms. Biochemistry (1998) 37(17):5961–7. doi:10.1021/bi971763a
179. Walter J, Fluhrer R, Hartung B, Willem M, Kaether C, Capell A, et al. Phosphorylation regulates intracellular trafficking of beta-secretase. J Biol Chem (2001) 276(18):14634–41. doi:10.1074/jbc.M011116200
180. Yamamoto A, Friedlein A, Imai Y, Takahashi R, Kahle PJ, Haass C. Parkin phosphorylation and modulation of its E3 ubiquitin ligase activity. J Biol Chem (2005) 280(5):3390–9. doi:10.1074/jbc.M407724200
181. Okochi M, Walter J, Koyama A, Nakajo S, Baba M, Iwatsubo T, et al. Constitutive phosphorylation of the Parkinson’s disease associated alpha-synuclein. J Biol Chem (2000) 275(1):390–7. doi:10.1074/jbc.275.1.390
182. Mishra SK, Yang Z, Mazumdar A, Talukder AH, Larose L, Kumar R. Metastatic tumor antigen 1 short form (MTA1s) associates with casein kinase I-gamma 2, an estrogen-responsive kinase. Oncogene (2004) 23(25):4422–9. doi:10.1038/sj.onc.1207569
183. Lohler J, Hirner H, Schmidt B, Kramer K, Fischer D, Thal DR, et al. Immunohistochemical characterisation of cell-type specific expression of CK1delta in various tissues of young adult BALB/c mice. PLoS One (2009) 4(1):e4174. doi:10.1371/journal.pone.0004174
184. Utz AC, Hirner H, Blatz A, Hillenbrand A, Schmidt B, Deppert W, et al. Analysis of cell type-specific expression of CK1 epsilon in various tissues of young adult BALB/c Mice and in mammary tumors of SV40 T-Ag-transgenic mice. J Histochem Cytochem (2010) 58(1):1–15. doi:10.1369/jhc.2009.954628
185. Cobb MH, Rosen OM. Description of a protein kinase derived from insulin-treated 3T3-L1 cells that catalyzes the phosphorylation of ribosomal protein S6 and casein. J Biol Chem (1983) 258(20):12472–81.
186. Elias L, Li AP, Longmire J. Cyclic adenosine 3′:5′-monophosphate-dependent and -independent protein kinase in acute myeloblastic leukemia. Cancer Res (1981) 41(6):2182–8.
187. Knippschild U, Milne DM, Campbell LE, DeMaggio AJ, Christenson E, Hoekstra MF, et al. p53 is phosphorylated in vitro and in vivo by the delta and epsilon isoforms of casein kinase 1 and enhances the level of casein kinase 1 delta in response to topoisomerase-directed drugs. Oncogene (1997) 15(14):1727–36. doi:10.1038/sj.onc.1201541
188. Santos JA, Logarinho E, Tapia C, Allende CC, Allende JE, Sunkel CE. The casein kinase 1 alpha gene of Drosophila melanogaster is developmentally regulated and the kinase activity of the protein induced by DNA damage. J Cell Sci (1996) 109(Pt 7):1847–56.
189. Deppert W, Kurth M, Graessmann M, Graessmann A, Knippschild U. Altered phosphorylation at specific sites confers a mutant phenotype to SV40 wild-type large T antigen in a flat revertant of SV40-transformed cells. Oncogene (1991) 6(10):1931–8.
190. Hirner H, Gunes C, Bischof J, Wolff S, Grothey A, Kuhl M, et al. Impaired CK1 delta activity attenuates SV40-induced cellular transformation in vitro and mouse mammary carcinogenesis in vivo. PLoS One (2012) 7(1):e29709. doi:10.1371/journal.pone.0029709
191. Sillibourne JE, Milne DM, Takahashi M, Ono Y, Meek DW. Centrosomal anchoring of the protein kinase CK1delta mediated by attachment to the large, coiled-coil scaffolding protein CG-NAP/AKAP450. J Mol Biol (2002) 322(4):785–97. doi:10.1016/S0022-2836(02)00857-4
192. Vancura A, Sessler A, Leichus B, Kuret J. A prenylation motif is required for plasma membrane localization and biochemical function of casein kinase I in budding yeast. J Biol Chem (1994) 269(30):19271–8.
193. Milne DM, Looby P, Meek DW. Catalytic activity of protein kinase CK1 delta (casein kinase 1 delta) is essential for its normal subcellular localization. Exp Cell Res (2001) 263(1):43–54. doi:10.1006/excr.2000.5100
194. Yin H, Laguna KA, Li G, Kuret J. Dysbindin structural homologue CK1BP is an isoform-selective binding partner of human casein kinase-1. Biochemistry (2006) 45(16):5297–308. doi:10.1021/bi052354e
195. Cheong JK, Virshup DM. Casein kinase 1: complexity in the family. Int J Biochem Cell Biol (2011) 43(4):465–9. doi:10.1016/j.biocel.2010.12.004
196. Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science (2011) 332(6030):680–6. doi:10.1126/science.1198701
197. Locasale JW, Shaw AS, Chakraborty AK. Scaffold proteins confer diverse regulatory properties to protein kinase cascades. Proc Natl Acad Sci U S A (2007) 104(33):13307–12. doi:10.1073/pnas.0706311104
198. Kholodenko BN, Hancock JF, Kolch W. Signalling ballet in space and time. Nat Rev Mol Cell Biol (2010) 11(6):414–26. doi:10.1038/nrm2901
199. Brown D, Hikim AP, Kovacheva EL, Sinha-Hikim I. Mouse model of testosterone-induced muscle fiber hypertrophy: involvement of p38 mitogen-activated protein kinase-mediated Notch signaling. J Endocrinol (2009) 201(1):129–39. doi:10.1677/JOE-08-0476
200. Gu L, Fullam A, Brennan R, Schroder M. Human DEAD box helicase 3 couples IkappaB kinase epsilon to interferon regulatory factor 3 activation. Mol Cell Biol (2013) 33(10):2004–15. doi:10.1128/MCB.01603-12
201. Greer YE, Westlake CJ, Gao B, Bharti K, Shiba Y, Xavier CP, et al. Casein kinase 1 delta functions at the centrosome and golgi to promote ciliogenesis. Mol Biol Cell (2014). doi:10.1091/mbc.E13-10-0598
202. Cegielska A, Gietzen KF, Rivers A, Virshup DM. Autoinhibition of casein kinase I epsilon (CKI epsilon) is relieved by protein phosphatases and limited proteolysis. J Biol Chem (1998) 273(3):1357–64. doi:10.1074/jbc.273.3.1357
203. Gietzen KF, Virshup DM. Identification of inhibitory autophosphorylation sites in casein kinase I epsilon. J Biol Chem (1999) 274(45):32063–70. doi:10.1074/jbc.274.45.32063
204. Graves PR, Roach PJ. Role of COOH-terminal phosphorylation in the regulation of casein kinase I delta. J Biol Chem (1995) 270(37):21689–94. doi:10.1074/jbc.270.37.21689
205. Rivers A, Gietzen KF, Vielhaber E, Virshup DM. Regulation of casein kinase I epsilon and casein kinase I delta by an in vivo futile phosphorylation cycle. J Biol Chem (1998) 273(26):15980–4. doi:10.1074/jbc.273.26.15980
206. Budini M, Jacob G, Jedlicki A, Perez C, Allende CC, Allende JE. Autophosphorylation of carboxy-terminal residues inhibits the activity of protein kinase CK1alpha. J Cell Biochem (2009) 106(3):399–408. doi:10.1002/jcb.22019
207. Bedri S, Cizek SM, Rastarhuyeva I, Stone JR. Regulation of protein kinase CK1alphaLS by dephosphorylation in response to hydrogen peroxide. Arch Biochem Biophys (2007) 466(2):242–9. doi:10.1016/j.abb.2007.06.010
208. Huart AS, MacLaine NJ, Narayan V, Hupp TR. Exploiting the MDM2-CK1alpha protein-protein interface to develop novel biologics that induce UBL-kinase-modification and inhibit cell growth. PLoS One (2012) 7(8):e43391. doi:10.1371/journal.pone.0043391
209. De Maio A. Heat shock proteins: facts, thoughts, and dreams. Shock (1999) 11(1):1–12. doi:10.1097/00024382-199901000-00001
210. Maritzen T, Lohler J, Deppert W, Knippschild U. Casein kinase I delta (CKIdelta) is involved in lymphocyte physiology. Eur J Cell Biol (2003) 82(7):369–78. doi:10.1078/0171-9335-00323
211. Shibasaki F, Price ER, Milan D, McKeon F. Role of kinases and the phosphatase calcineurin in the nuclear shuttling of transcription factor NF-AT4. Nature (1996) 382(6589):370–3. doi:10.1038/382370a0
212. Marin O, Burzio V, Boschetti M, Meggio F, Allende CC, Allende JE, et al. Structural features underlying the multisite phosphorylation of the A domain of the NF-AT4 transcription factor by protein kinase CK1. Biochemistry (2002) 41(2):618–27. doi:10.1021/bi0112309
213. Okamura H, Garcia-Rodriguez C, Martinson H, Qin J, Virshup DM, Rao A. A conserved docking motif for CK1 binding controls the nuclear localization of NFAT1. Mol Cell Biol (2004) 24(10):4184–95. doi:10.1128/MCB.24.10.4184-4195.2004
214. Dustin ML. T-cell activation through immunological synapses and kinapses. Immunol Rev (2008) 221:77–89. doi:10.1111/j.1600-065X.2008.00589.x
215. Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell (2012) 148(3):399–408. doi:10.1016/j.cell.2012.01.021
216. Kalousi A, Mylonis I, Politou AS, Chachami G, Paraskeva E, Simos G. Casein kinase 1 regulates human hypoxia-inducible factor HIF-1. J Cell Sci (2010) 123(Pt 17):2976–86. doi:10.1242/jcs.068122
217. Okamura A, Iwata N, Nagata A, Tamekane A, Shimoyama M, Gomyo H, et al. Involvement of casein kinase I epsilon in cytokine-induced granulocytic differentiation. Blood (2004) 103(8):2997–3004. doi:10.1182/blood-2003-08-2768
218. Brockman JL, Gross SD, Sussman MR, Anderson RA. Cell cycle-dependent localization of casein kinase I to mitotic spindles. Proc Natl Acad Sci U S A (1992) 89(20):9454–8. doi:10.1073/pnas.89.20.9454
219. Knippschild U, Gocht A, Wolff S, Huber N, Lohler J, Stoter M. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell Signal (2005) 17(6):675–89. doi:10.1016/j.cellsig.2004.12.011
220. Wang L, Lu A, Zhou HX, Sun R, Zhao J, Zhou CJ, et al. Casein kinase 1 alpha regulates chromosome congression and separation during mouse oocyte meiotic maturation and early embryo development. PLoS One (2013) 8(5):e63173. doi:10.1371/journal.pone.0063173
221. Behrend L, Milne DM, Stoter M, Deppert W, Campbell LE, Meek DW, et al. IC261, a specific inhibitor of the protein kinases casein kinase 1-delta and -epsilon, triggers the mitotic checkpoint and induces p53-dependent postmitotic effects. Oncogene (2000) 19(47):5303–13. doi:10.1038/sj.onc.1203939
222. Stoter M, Bamberger AM, Aslan B, Kurth M, Speidel D, Loning T, et al. Inhibition of casein kinase I delta alters mitotic spindle formation and induces apoptosis in trophoblast cells. Oncogene (2005) 24(54):7964–75. doi:10.1038/sj.onc.1208941
223. Ben-Nissan G, Cui W, Kim DJ, Yang Y, Yoo BC, Lee JY. Arabidopsis casein kinase 1-like 6 contains a microtubule-binding domain and affects the organization of cortical microtubules. Plant Physiol (2008) 148(4):1897–907. doi:10.1104/pp.108.129346
224. Drewes G, Ebneth A, Mandelkow EM. MAPs, MARKs and microtubule dynamics. Trends Biochem Sci (1998) 23(8):307–11. doi:10.1016/S0968-0004(98)01245-6
225. Hanger DP, Byers HL, Wray S, Leung KY, Saxton MJ, Seereeram A, et al. Novel phosphorylation sites in tau from Alzheimer brain support a role for casein kinase 1 in disease pathogenesis. J Biol Chem (2007) 282(32):23645–54. doi:10.1074/jbc.M703269200
226. Leon-Espinosa G, Garcia E, Garcia-Escudero V, Hernandez F, Defelipe J, Avila J. Changes in tau phosphorylation in hibernating rodents. J Neurosci Res (2013) 91(7):954–62. doi:10.1002/jnr.23220
227. Li G, Yin H, Kuret J. Casein kinase 1 delta phosphorylates tau and disrupts its binding to microtubules. J Biol Chem (2004) 279(16):15938–45. doi:10.1074/jbc.M314116200
228. Ikeda K, Zhapparova O, Brodsky I, Semenova I, Tirnauer JS, Zaliapin I, et al. CK1 activates minus-end-directed transport of membrane organelles along microtubules. Mol Biol Cell (2011) 22(8):1321–9. doi:10.1091/mbc.E10-09-0741
229. Greer YE, Rubin JS. Casein kinase 1 delta functions at the centrosome to mediate Wnt-3a-dependent neurite outgrowth. J Cell Biol (2011) 192(6):993–1004. doi:10.1083/jcb.201011111
230. Lee KH, Johmura Y, Yu LR, Park JE, Gao Y, Bang JK, et al. Identification of a novel Wnt5a-CK1 varepsilon-Dvl2-Plk1-mediated primary cilia disassembly pathway. EMBO J (2012) 31(14):3104–17. doi:10.1038/emboj.2012.144
231. Meek DW. The role of p53 in the response to mitotic spindle damage. Pathol Biol (Paris) (2000) 48(3):246–54.
232. Tarapore P, Fukasawa K. Loss of p53 and centrosome hyperamplification. Oncogene (2002) 21(40):6234–40. doi:10.1038/sj.onc.1205707
233. Wahl AF, Donaldson KL, Fairchild C, Lee FY, Foster SA, Demers GW, et al. Loss of normal p53 function confers sensitization to Taxol by increasing G2/M arrest and apoptosis. Nat Med (1996) 2(1):72–9. doi:10.1038/nm0196-72
234. Dumaz N, Milne DM, Meek DW. Protein kinase CK1 is a p53-threonine 18 kinase which requires prior phosphorylation of serine 15. FEBS Lett (1999) 463(3):312–6. doi:10.1016/S0014-5793(99)01647-6
235. Knippschild U, Milne D, Campbell L, Meek D. p53 N-terminus-targeted protein kinase activity is stimulated in response to wild type p53 and DNA damage. Oncogene (1996) 13(7):1387–93.
236. MacLaine NJ, Oster B, Bundgaard B, Fraser JA, Buckner C, Lazo PA, et al. A central role for CK1 in catalyzing phosphorylation of the p53 transactivation domain at serine 20 after HHV-6B viral infection. J Biol Chem (2008) 283(42):28563–73. doi:10.1074/jbc.M804433200
237. Venerando A, Marin O, Cozza G, Bustos VH, Sarno S, Pinna LA. Isoform specific phosphorylation of p53 by protein kinase CK1. Cell Mol Life Sci (2010) 67(7):1105–18. doi:10.1007/s00018-009-0236-7
238. Sakaguchi K, Saito S, Higashimoto Y, Roy S, Anderson CW, Appella E. Damage-mediated phosphorylation of human p53 threonine 18 through a cascade mediated by a casein 1-like kinase. Effect on Mdm2 binding. J Biol Chem (2000) 275(13):9278–83.
239. Kulikov R, Winter M, Blattner C. Binding of p53 to the central domain of Mdm2 is regulated by phosphorylation. J Biol Chem (2006) 281(39):28575–83. doi:10.1074/jbc.M513311200
240. Inuzuka H, Tseng A, Gao D, Zhai B, Zhang Q, Shaik S, et al. Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF(beta-TRCP) ubiquitin ligase. Cancer Cell (2010) 18(2):147–59. doi:10.1016/j.ccr.2010.06.015
241. Meek DW, Knippschild U. Posttranslational modification of MDM2. Mol Cancer Res (2003) 1(14):1017–26.
242. Huart AS, MacLaine NJ, Meek DW, Hupp TR. CK1alpha plays a central role in mediating MDM2 control of p53 and E2F-1 protein stability. J Biol Chem (2009) 284(47):32384–94. doi:10.1074/jbc.M109.052647
243. Hammond EM, Denko NC, Dorie MJ, Abraham RT, Giaccia AJ. Hypoxia links ATR and p53 through replication arrest. Mol Cell Biol (2002) 22(6):1834–43. doi:10.1128/MCB.22.6.1834-1843.2002
244. Hammond EM, Dorie MJ, Giaccia AJ. ATR/ATM targets are phosphorylated by ATR in response to hypoxia and ATM in response to reoxygenation. J Biol Chem (2003) 278(14):12207–13. doi:10.1074/jbc.M212360200
245. Takaoka A, Hayakawa S, Yanai H, Stoiber D, Negishi H, Kikuchi H, et al. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature (2003) 424(6948):516–23. doi:10.1038/nature01850
246. Leonova KI, Brodsky L, Lipchick B, Pal M, Novototskaya L, Chenchik AA, et al. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc Natl Acad Sci U S A (2013) 110(1):E89–98. doi:10.1073/pnas.1216922110
247. Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, et al. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature (2008) 455(7214):822–5. doi:10.1038/nature07273
248. Bargonetti J, Reynisdottir I, Friedman PN, Prives C. Site-specific binding of wild-type p53 to cellular DNA is inhibited by SV40 T antigen and mutant p53. Genes Dev (1992) 6(10):1886–98. doi:10.1101/gad.6.10.1886
249. Henning W, Rohaly G, Kolzau T, Knippschild U, Maacke H, Deppert W. MDM2 is a target of simian virus 40 in cellular transformation and during lytic infection. J Virol (1997) 71(10):7609–18.
250. Li L, Zhao D, Wei H, Yao L, Dang Y, Amjad A, et al. REGgamma deficiency promotes premature aging via the casein kinase 1 pathway. Proc Natl Acad Sci U S A (2013) 110(27):11005–10. doi:10.1073/pnas.1308497110
251. Iliakis G, Wang Y, Guan J, Wang H. DNA damage checkpoint control in cells exposed to ionizing radiation. Oncogene (2003) 22(37):5834–47. doi:10.1038/sj.onc.1206682
252. Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell (1993) 75(4):805–16. doi:10.1016/0092-8674(93)90499-G
253. Gottifredi V, Karni-Schmidt O, Shieh SS, Prives C. p53 down-regulates CHK1 through p21 and the retinoblastoma protein. Mol Cell Biol (2001) 21(4):1066–76. doi:10.1128/MCB.21.4.1066-1076.2001
254. Martelli AM, Tazzari PL, Tabellini G, Bortul R, Billi AM, Manzoli L, et al. A new selective AKT pharmacological inhibitor reduces resistance to chemotherapeutic drugs, TRAIL, all-trans-retinoic acid, and ionizing radiation of human leukemia cells. Leukemia (2003) 17(9):1794–805. doi:10.1038/sj.leu.2403044
255. Pommier Y, Sordet O, Antony S, Hayward RL, Kohn KW. Apoptosis defects and chemotherapy resistance: molecular interaction maps and networks. Oncogene (2004) 23(16):2934–49. doi:10.1038/sj.onc.1207515
256. Okamura A, Iwata N, Tamekane A, Yakushijin K, Nishikawa S, Hamaguchi M, et al. Casein kinase Iepsilon down-regulates phospho-Akt via PTEN, following genotoxic stress-induced apoptosis in hematopoietic cells. Life Sci (2006) 78(14):1624–9. doi:10.1016/j.lfs.2005.07.041
257. Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol (2005) 17(6):596–603. doi:10.1016/j.ceb.2005.09.009
258. Duan S, Skaar JR, Kuchay S, Toschi A, Kanarek N, Ben-Neriah Y, et al. mTOR generates an auto-amplification loop by triggering the betaTrCP- and CK1alpha-dependent degradation of DEPTOR. Mol Cell (2011) 44(2):317–24. doi:10.1016/j.molcel.2011.09.005
259. Carrasco DR, Tonon G, Huang Y, Zhang Y, Sinha R, Feng B, et al. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell (2006) 9(4):313–25. doi:10.1016/j.ccr.2006.03.019
260. Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell (2009) 137(5):873–86. doi:10.1016/j.cell.2009.03.046
261. Roh DS, Funderburgh JL. Rapid changes in connexin-43 in response to genotoxic stress stabilize cell-cell communication in corneal endothelium. Invest Ophthalmol Vis Sci (2011) 52(8):5174–82. doi:10.1167/iovs.11-7272
262. Kelleher FC, Rao A, Maguire A. Circadian molecular clocks and cancer. Cancer Lett (2014) 342(1):9–18. doi:10.1016/j.canlet.2013.09.040
263. Kategaya LS, Hilliard A, Zhang L, Asara JM, Ptacek LJ, Fu YH. Casein kinase 1 proteomics reveal prohibitin 2 function in molecular clock. PLoS One (2012) 7(2):e31987. doi:10.1371/journal.pone.0031987
264. Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell (2006) 22(3):375–82. doi:10.1016/j.molcel.2006.03.038
265. Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol (2005) 25(8):3109–16. doi:10.1128/MCB.25.8.3109-3116.2005
266. Mullenders J, Fabius AW, Madiredjo M, Bernards R, Beijersbergen RL. A large scale shRNA barcode screen identifies the circadian clock component ARNTL as putative regulator of the p53 tumor suppressor pathway. PLoS One (2009) 4(3):e4798. doi:10.1371/journal.pone.0004798
267. Roberts CK, Sindhu KK. Oxidative stress and metabolic syndrome. Life Sci (2009) 84(21–22):705–12. doi:10.1016/j.lfs.2009.02.026
268. Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev (2006) 20(14):1868–73. doi:10.1101/gad.1432206
269. Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science (2001) 293(5529):510–4. doi:10.1126/science.1060698
270. Wood PA, Yang X, Taber A, Oh EY, Ansell C, Ayers SE, et al. Period 2 mutation accelerates ApcMin/+ tumorigenesis. Mol Cancer Res (2008) 6(11):1786–93. doi:10.1158/1541-7786.MCR-08-0196
271. Virshup DM, Eide EJ, Forger DB, Gallego M, Harnish EV. Reversible protein phosphorylation regulates circadian rhythms. Cold Spring Harb Symp Quant Biol (2007) 72:413–20. doi:10.1101/sqb.2007.72.048
272. Etchegaray JP, Machida KK, Noton E, Constance CM, Dallmann R, Di Napoli MN, et al. Casein kinase 1 delta regulates the pace of the mammalian circadian clock. Mol Cell Biol (2009) 29(14):3853–66. doi:10.1128/MCB.00338-09
273. Walton KM, Fisher K, Rubitski D, Marconi M, Meng QJ, Sladek M, et al. Selective inhibition of casein kinase 1 epsilon minimally alters circadian clock period. J Pharmacol Exp Ther (2009) 330(2):430–9. doi:10.1124/jpet.109.151415
274. Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell (2001) 107(7):855–67. doi:10.1016/S0092-8674(01)00610-9
275. Hirota T, Lee JW, Lewis WG, Zhang EE, Breton G, Liu X, et al. High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIalpha as a clock regulatory kinase. PLoS Biol (2010) 8(12):e1000559. doi:10.1371/journal.pbio.1000559
276. Lee HM, Chen R, Kim H, Etchegaray JP, Weaver DR, Lee C. The period of the circadian oscillator is primarily determined by the balance between casein kinase 1 and protein phosphatase 1. Proc Natl Acad Sci U S A (2011) 108(39):16451–6. doi:10.1073/pnas.1107178108
277. Isojima Y, Nakajima M, Ukai H, Fujishima H, Yamada RG, Masumoto KH, et al. CKIepsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc Natl Acad Sci U S A (2009) 106(37):15744–9. doi:10.1073/pnas.0908733106
278. Meng QJ, Maywood ES, Bechtold DA, Lu WQ, Li J, Gibbs JE, et al. Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc Natl Acad Sci U S A (2010) 107(34):15240–5. doi:10.1073/pnas.1005101107
279. Maywood ES, Chesham JE, Smyllie NJ, Hastings MH. The Tau mutation of casein kinase 1{epsilon} sets the period of the mammalian pacemaker via regulation of period1 or period2 clock proteins. J Biol Rhythms (2014) 29(2):110–8. doi:10.1177/0748730414520663
280. Meng QJ, Logunova L, Maywood ES, Gallego M, Lebiecki J, Brown TM, et al. Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron (2008) 58(1):78–88. doi:10.1016/j.neuron.2008.01.019
281. Um JH, Yang S, Yamazaki S, Kang H, Viollet B, Foretz M, et al. Activation of 5′-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)-dependent degradation of clock protein mPer2. J Biol Chem (2007) 282(29):20794–8. doi:10.1074/jbc.C700070200
282. Partch CL, Shields KF, Thompson CL, Selby CP, Sancar A. Posttranslational regulation of the mammalian circadian clock by cryptochrome and protein phosphatase 5. Proc Natl Acad Sci U S A (2006) 103(27):10467–72. doi:10.1073/pnas.0604138103
283. Storz S, Tovin A, Mracek P, Alon S, Foulkes N, Gothilf Y. Casein kinase 1 delta activity: a key element in the Zebrafish circadian timing system. PLoS One (2013) 8(1):e54189. doi:10.1371/journal.pone.0054189
284. Beyaert R, Vanhaesebroeck B, Declercq W, Van Lint J, Vandenabele P, Agostinis P, et al. Casein kinase-1 phosphorylates the p75 tumor necrosis factor receptor and negatively regulates tumor necrosis factor signaling for apoptosis. J Biol Chem (1995) 270(40):23293–9. doi:10.1074/jbc.270.40.23293
285. Katagiri Y, Takeda K, Yu ZX, Ferrans VJ, Ozato K, Guroff G. Modulation of retinoid signalling through NGF-induced nuclear export of NGFI-B. Nat Cell Biol (2000) 2(7):435–40. doi:10.1038/35017072
286. Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev (2000) 14(12):1448–59.
287. Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell (1995) 83(6):841–50. doi:10.1016/0092-8674(95)90200-7
288. Alappat EC, Volkland J, Peter ME. Cell cycle effects by C-FADD depend on its C-terminal phosphorylation site. J Biol Chem (2003) 278(43):41585–8. doi:10.1074/jbc.C300385200
289. Zelenak C, Eberhard M, Jilani K, Qadri SM, Macek B, Lang F. Protein kinase CK1alpha regulates erythrocyte survival. Cell Physiol Biochem (2012) 29(1–2):171–80. doi:10.1159/000337598
290. Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev (2001) 15(23):3059–87. doi:10.1101/gad.938601
291. Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol (2004) 20:781–810. doi:10.1146/annurev.cellbio.20.010403.113126
292. Pan D. Hippo signaling in organ size control. Genes Dev (2007) 21(8):886–97. doi:10.1101/gad.1536007
293. Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol (2011) 13(8):877–83. doi:10.1038/ncb2303
294. Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer (2008) 8(5):387–98. doi:10.1038/nrc2389
295. Bao Y, Hata Y, Ikeda M, Withanage K. Mammalian Hippo pathway: from development to cancer and beyond. J Biochem (2011) 149(4):361–79. doi:10.1093/jb/mvr021
296. Clevers H. Wnt/beta-catenin signaling in development and disease. Cell (2006) 127(3):469–80. doi:10.1016/j.cell.2006.10.018
297. Pan D. The hippo signaling pathway in development and cancer. Dev Cell (2010) 19(4):491–505. doi:10.1016/j.devcel.2010.09.011
298. Rubin LL, de Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nat Rev Drug Discov (2006) 5(12):1026–33. doi:10.1038/nrd2086
299. Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev (1997) 11(24):3286–305. doi:10.1101/gad.11.24.3286
300. McMahon AP, Moon RT. Ectopic expression of the proto-oncogene int-1 in Xenopus embryos leads to duplication of the embryonic axis. Cell (1989) 58(6):1075–84. doi:10.1016/0092-8674(89)90506-0
301. Moon RT, Brown JD, Torres M. WNTs modulate cell fate and behavior during vertebrate development. Trends Genet (1997) 13(4):157–62. doi:10.1016/S0168-9525(97)01093-7
302. Rocheleau CE, Downs WD, Lin R, Wittmann C, Bei Y, Cha YH, et al. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell (1997) 90(4):707–16. doi:10.1016/S0092-8674(00)80531-0
303. Chan EF, Gat U, McNiff JM, Fuchs E. A common human skin tumour is caused by activating mutations in beta-catenin. Nat Genet (1999) 21(4):410–3. doi:10.1038/7747
304. Koch A, Denkhaus D, Albrecht S, Leuschner I, von Schweinitz D, Pietsch T. Childhood hepatoblastomas frequently carry a mutated degradation targeting box of the beta-catenin gene. Cancer Res (1999) 59(2):269–73.
305. Nishisho I, Nakamura Y, Miyoshi Y, Miki Y, Ando H, Horii A, et al. Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science (1991) 253(5020):665–9. doi:10.1126/science.1651563
307. Bass AJ, Lawrence MS, Brace LE, Ramos AH, Drier Y, Cibulskis K, et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet (2011) 43(10):964–8. doi:10.1038/ng.936
308. Kinzler KW, Nilbert MC, Su LK, Vogelstein B, Bryan TM, Levy DB, et al. Identification of FAP locus genes from chromosome 5q21. Science (1991) 253(5020):661–5. doi:10.1126/science.1651562
309. Liu W, Dong X, Mai M, Seelan RS, Taniguchi K, Krishnadath KK, et al. Mutations in AXIN2 cause colorectal cancer with defective mismatch repair by activating beta-catenin/TCF signalling. Nat Genet (2000) 26(2):146–7. doi:10.1038/79859
310. Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science (1997) 275(5307):1787–90. doi:10.1126/science.275.5307.1787
311. Rubinfeld B, Souza B, Albert I, Muller O, Chamberlain SH, Masiarz FR, et al. Association of the APC gene product with beta-catenin. Science (1993) 262(5140):1731–4. doi:10.1126/science.8259518
312. Takeda H, Lyle S, Lazar AJ, Zouboulis CC, Smyth I, Watt FM. Human sebaceous tumors harbor inactivating mutations in LEF1. Nat Med (2006) 12(4):395–7. doi:10.1038/nm1386
313. Umar S, Wang Y, Morris AP, Sellin JH. Dual alterations in casein kinase I-epsilon and GSK-3beta modulate beta-catenin stability in hyperproliferating colonic epithelia. Am J Physiol Gastrointest Liver Physiol (2007) 292(2):G599–607. doi:10.1152/ajpgi.00343.2006
314. Swiatek W, Kang H, Garcia BA, Shabanowitz J, Coombs GS, Hunt DF, et al. Negative regulation of LRP6 function by casein kinase I epsilon phosphorylation. J Biol Chem (2006) 281(18):12233–41. doi:10.1074/jbc.M510580200
315. Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature (2004) 432(7015):324–31. doi:10.1038/nature03100
316. Wicking C, Smyth I, Bale A. The hedgehog signalling pathway in tumorigenesis and development. Oncogene (1999) 18(55):7844–51. doi:10.1038/sj.onc.1203282
317. Lindemann RK. Stroma-initiated hedgehog signaling takes center stage in B-cell lymphoma. Cancer Res (2008) 68(4):961–4. doi:10.1158/0008-5472.CAN-07-5500
318. Ruiz i Altaba A, Mas C, Stecca B. The Gli code: an information nexus regulating cell fate, stemness and cancer. Trends Cell Biol (2007) 17(9):438–47. doi:10.1016/j.tcb.2007.06.007
319. Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes Dev (2008) 22(18):2454–72. doi:10.1101/gad.1693608
320. Denef N, Neubuser D, Perez L, Cohen SM. Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell (2000) 102(4):521–31. doi:10.1016/S0092-8674(00)00056-8
321. Taipale J, Cooper MK, Maiti T, Beachy PA. Patched acts catalytically to suppress the activity of Smoothened. Nature (2002) 418(6900):892–7. doi:10.1038/nature00989
322. Price MA, Kalderon D. Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by Glycogen Synthase Kinase 3 and Casein Kinase 1. Cell (2002) 108(6):823–35. doi:10.1016/S0092-8674(02)00664-5
323. Price MA. CKI, there’s more than one: casein kinase I family members in Wnt and Hedgehog signaling. Genes Dev (2006) 20(4):399–410. doi:10.1101/gad.1394306
324. Aza-Blanc P, Lin HY, Ruiz i Altaba A, Kornberg TB. Expression of the vertebrate Gli proteins in Drosophila reveals a distribution of activator and repressor activities. Development (2000) 127(19):4293–301.
325. Hammerschmidt M, Bitgood MJ, McMahon AP. Protein kinase A is a common negative regulator of Hedgehog signaling in the vertebrate embryo. Genes Dev (1996) 10(6):647–58. doi:10.1101/gad.10.6.647
326. von Mering C, Basler K. Distinct and regulated activities of human Gli proteins in Drosophila. Curr Biol (1999) 9(22):1319–22. doi:10.1016/S0960-9822(00)80054-8
327. Wang B, Li Y. Evidence for the direct involvement of {beta}TrCP in Gli3 protein processing. Proc Natl Acad Sci U S A (2006) 103(1):33–8. doi:10.1073/pnas.0509927103
328. Chen Y, Sasai N, Ma G, Yue T, Jia J, Briscoe J, et al. Sonic Hedgehog dependent phosphorylation by CK1alpha and GRK2 is required for ciliary accumulation and activation of smoothened. PLoS Biol (2011) 9(6):e1001083. doi:10.1371/journal.pbio.1001083
329. Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer (2013) 13(4):246–57. doi:10.1038/nrc3458
330. Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell (2007) 130(6):1120–33. doi:10.1016/j.cell.2007.07.019
331. Harvey K, Tapon N. The Salvador-Warts-Hippo pathway – an emerging tumour-suppressor network. Nat Rev Cancer (2007) 7(3):182–91. doi:10.1038/nrc2070
332. Xu F, Wang YL, Chang JJ, Du SC, Diao L, Jiang N, et al. Mammalian sterile 20-like kinase 1/2 inhibits the Wnt/beta-catenin signalling pathway by directly binding casein kinase 1epsilon. Biochem J (2014) 458(1):159–69. doi:10.1042/BJ20130986
333. Knippschild U, Wolff S, Giamas G, Brockschmidt C, Wittau M, Wurl PU, et al. The role of the casein kinase 1 (CK1) family in different signaling pathways linked to cancer development. Onkologie (2005) 28(10):508–14. doi:10.1159/000087137
334. Foldynova-Trantirkova S, Sekyrova P, Tmejova K, Brumovska E, Bernatik O, Blankenfeldt W, et al. Breast cancer-specific mutations in CK1 epsilon inhibit Wnt/beta-catenin and activate the Wnt/Rac1/JNK and NFAT pathways to decrease cell adhesion and promote cell migration. Breast Cancer Res (2010) 12(3):R30. doi:10.1186/bcr2581
335. Tsai IC, Woolf M, Neklason DW, Branford WW, Yost HJ, Burt RW, et al. Disease-associated casein kinase I delta mutation may promote adenomatous polyps formation via a Wnt/beta-catenin independent mechanism. Int J Cancer (2007) 120(5):1005–12. doi:10.1002/ijc.22368
336. Elyada E, Pribluda A, Goldstein RE, Morgenstern Y, Brachya G, Cojocaru G, et al. CKIalpha ablation highlights a critical role for p53 in invasiveness control. Nature (2011) 470(7334):409–13. doi:10.1038/nature09673
337. Sinnberg T, Menzel M, Kaesler S, Biedermann T, Sauer B, Nahnsen S, et al. Suppression of casein kinase 1alpha in melanoma cells induces a switch in beta-catenin signaling to promote metastasis. Cancer Res (2010) 70(17):6999–7009. doi:10.1158/0008-5472.CAN-10-0645
338. Masuda K, Ono M, Okamoto M, Morikawa W, Otsubo M, Migita T, et al. Downregulation of Cap43 gene by von Hippel-Lindau tumor suppressor protein in human renal cancer cells. Int J Cancer (2003) 105(6):803–10. doi:10.1002/ijc.11152
339. Brockschmidt C, Hirner H, Huber N, Eismann T, Hillenbrand A, Giamas G, et al. Anti-apoptotic and growth-stimulatory functions of CK1 delta and epsilon in ductal adenocarcinoma of the pancreas are inhibited by IC261 in vitro and in vivo. Gut (2008) 57(6):799–806. doi:10.1136/gut.2007.123695
340. Relles D, Sendecki J, Chipitsyna G, Hyslop T, Yeo CJ, Arafat HA. Circadian gene expression and clinicopathologic correlates in pancreatic cancer. J Gastrointest Surg (2013) 17(3):443–50. doi:10.1007/s11605-012-2112-2
341. Fuja TJ, Lin F, Osann KE, Bryant PJ. Somatic mutations and altered expression of the candidate tumor suppressors CSNK1 epsilon, DLG1, and EDD/hHYD in mammary ductal carcinoma. Cancer Res (2004) 64(3):942–51. doi:10.1158/0008-5472.CAN-03-2100
342. Shin S, Wolgamott L, Roux PP, Yoon SO. Casein Kinase 1{varepsilon} Promotes Cell Proliferation by Regulating mRNA Translation. Cancer Res (2014) 74(1):201–11. doi:10.1002/jcb.23247
343. Frierson HF Jr., El-Naggar AK, Welsh JB, Sapinoso LM, Su AI, Cheng J, et al. Large scale molecular analysis identifies genes with altered expression in salivary adenoid cystic carcinoma. Am J Pathol (2002) 161(4):1315–23. doi:10.1016/S0002-9440(10)64408-2
344. Rodriguez N, Yang J, Hasselblatt K, Liu S, Zhou Y, Rauh-Hain JA, et al. Casein kinase I epsilon interacts with mitochondrial proteins for the growth and survival of human ovarian cancer cells. EMBO Mol Med (2012) 4(9):952–63. doi:10.1002/emmm.201101094
345. Yang WS, Stockwell BR. Inhibition of casein kinase 1-epsilon induces cancer-cell-selective, PERIOD2-dependent growth arrest. Genome Biol (2008) 9(6):R92. doi:10.1186/gb-2008-9-6-r92
346. Toyoshima M, Howie HL, Imakura M, Walsh RM, Annis JE, Chang AN, et al. Functional genomics identifies therapeutic targets for MYC-driven cancer. Proc Natl Acad Sci U S A (2012) 109(24):9545–50. doi:10.1073/pnas.1121119109
347. Lin SH, Lin YM, Yeh CM, Chen CJ, Chen MW, Hung HF, et al. Casein kinase 1 epsilon predicts poorer prognosis in low T-stage oral cancer patients. Int J Mol Sci (2014) 15:2876–91. doi:10.3390/ijms15022876
348. Jaras M, Miller PG, Chu LP, Puram RV, Fink EC, Schneider RK, et al. Csnk1a1 inhibition has p53-dependent therapeutic efficacy in acute myeloid leukemia. J Exp Med (2014) 211(4):605–12. doi:10.1084/jem.20131033
349. D’Angelo A, Zollo M. Unraveling genes and pathways influenced by H-prune PDE overexpression: a model to study cellular motility. Cell Cycle (2004) 3(6):758–61. doi:10.4161/cc.3.6.926
350. Jenkins Y, Markovtsov V, Lang W, Sharma P, Pearsall D, Warner J, et al. Critical role of the ubiquitin ligase activity of UHRF1, a nuclear RING finger protein, in tumor cell growth. Mol Biol Cell (2005) 16(12):5621–9. doi:10.1091/mbc.E05-03-0194
351. Wang F, Yang YZ, Shi CZ, Zhang P, Moyer MP, Zhang HZ, et al. UHRF1 promotes cell growth and metastasis through repression of p16(ink(4)a) in colorectal cancer. Ann Surg Oncol (2012) 19(8):2753–62. doi:10.1245/s10434-011-2194-1
352. Lee YG, Macoska JA, Korenchuk S, Pienta KJ. MIM, a potential metastasis suppressor gene in bladder cancer. Neoplasia (2002) 4(4):291–4. doi:10.1038/sj.neo.7900231
353. Medrek C, Landberg G, Andersson T, Leandersson K. Wnt-5a-CKI{alpha} signaling promotes {beta}-catenin/E-cadherin complex formation and intercellular adhesion in human breast epithelial cells. J Biol Chem (2009) 284(16):10968–79. doi:10.1074/jbc.M804923200
354. Jonsson M, Andersson T. Repression of Wnt-5a impairs DDR1 phosphorylation and modifies adhesion and migration of mammary cells. J Cell Sci (2001) 114(Pt 11):2043–53.
355. Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol (2000) 2(2):84–9. doi:10.1038/35000034
356. Chijiwa T, Hagiwara M, Hidaka H. A newly synthesized selective casein kinase I inhibitor, N-(2-aminoethyl)-5-chloroisoquinoline-8-sulfonamide, and affinity purification of casein kinase I from bovine testis. J Biol Chem (1989) 264(9):4924–7.
357. Rena G, Bain J, Elliott M, Cohen P. D4476, a cell-permeant inhibitor of CK1, suppresses the site-specific phosphorylation and nuclear exclusion of FOXO1a. EMBO Rep (2004) 5(1):60–5. doi:10.1038/sj.embor.7400048
358. Mashhoon N, DeMaggio AJ, Tereshko V, Bergmeier SC, Egli M, Hoekstra MF, et al. Crystal structure of a conformation-selective casein kinase-1 inhibitor. J Biol Chem (2000) 275(26):20052–60. doi:10.1074/jbc.M001713200
359. Badura L, Swanson T, Adamowicz W, Adams J, Cianfrogna J, Fisher K, et al. An inhibitor of casein kinase I epsilon induces phase delays in circadian rhythms under free-running and entrained conditions. J Pharmacol Exp Ther (2007) 322(2):730–8. doi:10.1124/jpet.107.122846
360. Oumata N, Bettayeb K, Ferandin Y, Demange L, Lopez-Giral A, Goddard ML, et al. Roscovitine-derived, dual-specificity inhibitors of cyclin-dependent kinases and casein kinases 1. J Med Chem (2008) 51(17):5229–42. doi:10.1021/jm800109e
361. Andrzejewska M, Pagano MA, Meggio F, Brunati AM, Kazimierczuk Z. Polyhalogenobenzimidazoles: synthesis and their inhibitory activity against casein kinases. Bioorg Med Chem (2003) 11(18):3997–4002. doi:10.1016/S0968-0896(03)00403-6
362. Cozza G, Gianoncelli A, Montopoli M, Caparrotta L, Venerando A, Meggio F, et al. Identification of novel protein kinase CK1 delta (CK1delta) inhibitors through structure-based virtual screening. Bioorg Med Chem Lett (2008) 18(20):5672–5. doi:10.1016/j.bmcl.2008.08.072
363. Yokoyama T, Okano M, Noshita T, Funayama S, Ohtsuki K. Characterization of (-)-matairesinol as a potent inhibitor of casein kinase I in vitro. Biol Pharm Bull (2003) 26(3):371–4. doi:10.1248/bpb.26.371
364. Baunbaek D, Trinkler N, Ferandin Y, Lozach O, Ploypradith P, Rucirawat S, et al. Anticancer alkaloid lamellarins inhibit protein kinases. Mar Drugs (2008) 6(4):514–27. doi:10.3390/md20080026
365. Shanware NP, Williams LM, Bowler MJ, Tibbetts RS. Non-specific in vivo inhibition of CK1 by the pyridinyl imidazole p38 inhibitors SB 203580 and SB 202190. BMB Rep (2009) 42(3):142–7. doi:10.5483/BMBRep.2009.42.3.142
366. Bibian M, Rahaim RJ, Choi JY, Noguchi Y, Schurer S, Chen W, et al. Development of highly selective casein kinase 1 delta/1epsilon (CK1delta/epsilon) inhibitors with potent antiproliferative properties. Bioorg Med Chem Lett (2013) 23(15):4374–80. doi:10.1016/j.bmcl.2013.05.075
367. Bischof J, Leban J, Zaja M, Grothey A, Radunsky B, Othersen O, et al. 2-Benzamido-N-(1H-benzo[d]imidazol-2-yl)thiazole-4-carboxamide derivatives as potent inhibitors of CK1delta/epsilon. Amino Acids (2012) 43(4):1577–91. doi:10.1007/s00726-012-1234-x
368. Hua Z, Huang X, Bregman H, Chakka N, DiMauro EF, Doherty EM, et al. 2-Phenylamino-6-cyano-1H-benzimidazole-based isoform selective casein kinase 1 gamma (CK1gamma) inhibitors. Bioorg Med Chem Lett (2012) 22(17):5392–5. doi:10.1016/j.bmcl.2012.07.046
369. Yang LL, Li GB, Yan HX, Sun QZ, Ma S, Ji P, et al. Discovery of N6-phenyl-1H-pyrazolo[3,4-d]pyrimidine-3,6-diamine derivatives as novel CK1 inhibitors using common-feature pharmacophore model based virtual screening and hit-to-lead optimization. Eur J Med Chem (2012) 56:30–8. doi:10.1016/j.ejmech.2012.08.007
370. Arey R, McClung CA. An inhibitor of casein kinase 1 epsilon/delta partially normalizes the manic-like behaviors of the ClockDelta19 mouse. Behav Pharmacol (2012) 23(4):392–6. doi:10.1097/FBP.0b013e32835651fd
371. Huart AS, Saxty B, Merritt A, Nekulova M, Lewis S, Huang Y, et al. A Casein kinase 1/Checkpoint kinase 1 pyrazolo-pyridine protein kinase inhibitor as novel activator of the p53 pathway. Bioorg Med Chem Lett (2013) 23(20):5578–85. doi:10.1016/j.bmcl.2013.08.046
372. Muraki M, Ohkawara B, Hosoya T, Onogi H, Koizumi J, Koizumi T, et al. Manipulation of alternative splicing by a newly developed inhibitor of Clks. J Biol Chem (2004) 279(23):24246–54. doi:10.1074/jbc.M314298200
373. Salado IG, Redondo M, Bello ML, Perez C, Liachko NF, Kraemer BC, et al. Protein kinase CK-1 inhibitors as new potential drugs for amyotrophic lateral sclerosis. J Med Chem (2014) 57(6):2755–72. doi:10.1021/jm500065f
374. Sprouse J, Reynolds L, Kleiman R, Tate B, Swanson TA, Pickard GE. Chronic treatment with a selective inhibitor of casein kinase I delta/epsilon yields cumulative phase delays in circadian rhythms. Psychopharmacology (Berl) (2010) 210(4):569–76. doi:10.1007/s00213-010-1860-5
375. Sprouse J, Reynolds L, Swanson TA, Engwall M. Inhibition of casein kinase I epsilon/delta produces phase shifts in the circadian rhythms of Cynomolgus monkeys. Psychopharmacology (Berl) (2009) 204(4):735–42. doi:10.1007/s00213-009-1503-x
376. Li D, Herrera S, Bubula N, Nikitina E, Palmer AA, Hanck DA, et al. Casein kinase 1 enables nucleus accumbens amphetamine-induced locomotion by regulating AMPA receptor phosphorylation. J Neurochem (2011) 118(2):237–47. doi:10.1111/j.1471-4159.2011.07308.x
377. Bryant CD, Parker CC, Zhou L, Olker C, Chandrasekaran RY, Wager TT, et al. Csnk1e is a genetic regulator of sensitivity to psychostimulants and opioids. Neuropsychopharmacology (2012) 37(4):1026–35. doi:10.1038/npp.2011.287
378. Perreau-Lenz S, Vengeliene V, Noori HR, Merlo-Pich EV, Corsi MA, Corti C, et al. Inhibition of the casein-kinase-1-epsilon/delta prevents relapse-like alcohol drinking. Neuropsychopharmacology (2012) 37(9):2121–31. doi:10.1038/npp.2012.62
379. Kurihara T, Sakurai E, Toyomoto M, Kii I, Kawamoto D, Asada T, et al. Alleviation of behavioral hypersensitivity in mouse models of inflammatory pain with two structurally different casein kinase 1 (CK1) inhibitors. Mol Pain (2014) 10(1):17. doi:10.1186/1744-8069-10-17
380. Cheong JK, Nguyen TH, Wang H, Tan P, Voorhoeve PM, Lee SH, et al. IC261 induces cell cycle arrest and apoptosis of human cancer cells via CK1delta/varepsilon and Wnt/beta-catenin independent inhibition of mitotic spindle formation. Oncogene (2011) 30(22):2558–69. doi:10.1038/onc.2010.627
381. Hottecke N, Liebeck M, Baumann K, Schubenel R, Winkler E, Steiner H, et al. Inhibition of gamma-secretase by the CK1 inhibitor IC261 does not depend on CK1delta. Bioorg Med Chem Lett (2010) 20(9):2958–63. doi:10.1016/j.bmcl.2010.02.110
382. Perez DI, Gil C, Martinez A. Protein kinases CK1 and CK2 as new targets for neurodegenerative diseases. Med Res Rev (2011) 31(6):924–54. doi:10.1002/med.20207
383. Yasojima K, Kuret J, DeMaggio AJ, McGeer E, McGeer PL. Casein kinase 1 delta mRNA is upregulated in Alzheimer disease brain. Brain Res (2000) 865(1):116–20. doi:10.1016/S0006-8993(00)02200-9
384. Kuret J, Johnson GS, Cha D, Christenson ER, DeMaggio AJ, Hoekstra MF. Casein kinase 1 is tightly associated with paired-helical filaments isolated from Alzheimer’s disease brain. J Neurochem (1997) 69(6):2506–15. doi:10.1046/j.1471-4159.1997.69062506.x
385. Schwab C, DeMaggio AJ, Ghoshal N, Binder LI, Kuret J, McGeer PL. Casein kinase 1 delta is associated with pathological accumulation of tau in several neurodegenerative diseases. Neurobiol Aging (2000) 21(4):503–10. doi:10.1016/S0197-4580(00)00110-X
386. Flajolet M, He G, Heiman M, Lin A, Nairn AC, Greengard P. Regulation of Alzheimer’s disease amyloid-beta formation by casein kinase I. Proc Natl Acad Sci U S A (2007) 104(10):4159–64. doi:10.1073/pnas.0611236104
387. Chauhan A, Chauhan VP, Murakami N, Brockerhoff H, Wisniewski HM. Amyloid beta-protein stimulates casein kinase I and casein kinase II activities. Brain Res (1993) 629(1):47–52. doi:10.1016/0006-8993(93)90479-7
Keywords: casein kinase 1, cellular stress, centrosome, p53, signal transduction, tumorigenesis, inhibitor, disease
Citation: Knippschild U, Krüger M, Richter J, Xu P, García-Reyes B, Peifer C, Halekotte J, Bakulev V and Bischof J (2014) The CK1 family: contribution to cellular stress response and its role in carcinogenesis. Front. Oncol. 4:96. doi: 10.3389/fonc.2014.00096
Received: 27 February 2014; Accepted: 18 April 2014;
Published online: 19 May 2014.
Edited by:
Daniel Speidel, Children’s Medical Research Institute, AustraliaReviewed by:
Lisa Bouchier-Hayes, Baylor College of Medicine, USADavid Virshup, Duke-NUS Graduate Medical School Singapore, Singapore
Copyright: © 2014 Knippschild, Krüger, Richter, Xu, García-Reyes, Peifer, Halekotte, Bakulev and Bischof. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Uwe Knippschild, Department of General and Visceral Surgery, Surgery Centre, Ulm University Hospital, Albert-Einstein-Allee 23, 89081 Ulm, Germany e-mail: uwe.knippschild@uniklinik-ulm.de
 Uwe Knippschild
Uwe Knippschild Marc Krüger
Marc Krüger Julia Richter
Julia Richter Pengfei Xu
Pengfei Xu Balbina García-Reyes
Balbina García-Reyes Christian Peifer2
Christian Peifer2 Jakob Halekotte
Jakob Halekotte Joachim Bischof
Joachim Bischof